Gene Page: FMR1
Summary ?
| GeneID | 2332 |
| Symbol | FMR1 |
| Synonyms | FMRP|FRAXA|POF|POF1 |
| Description | fragile X mental retardation 1 |
| Reference | MIM:309550|HGNC:HGNC:3775|Ensembl:ENSG00000102081|HPRD:02398|Vega:OTTHUMG00000022606 |
| Gene type | protein-coding |
| Map location | Xq27.3 |
| Sherlock p-value | 0.675 |
| Fetal beta | -0.109 |
| Support | Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias,schizotypal | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
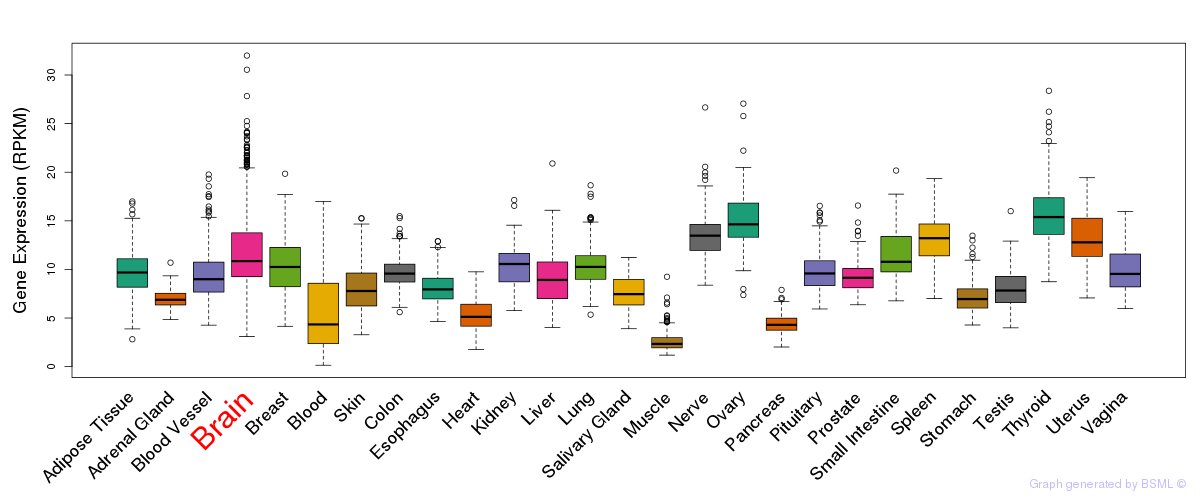
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CP | 0.91 | 0.50 |
| KCNJ13 | 0.90 | 0.41 |
| CLIC6 | 0.90 | 0.45 |
| TTR | 0.89 | 0.54 |
| F5 | 0.88 | 0.33 |
| SLC13A4 | 0.88 | 0.27 |
| PRLR | 0.85 | 0.20 |
| SLC5A5 | 0.85 | 0.24 |
| ABCA4 | 0.84 | 0.30 |
| TRPV4 | 0.82 | 0.31 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| EPHB6 | -0.17 | -0.36 |
| IER5L | -0.16 | -0.53 |
| CIDEA | -0.15 | -0.49 |
| RND1 | -0.15 | -0.37 |
| ARL4D | -0.15 | -0.48 |
| LNX1 | -0.14 | -0.41 |
| CACNB3 | -0.14 | -0.38 |
| MEF2C | -0.14 | -0.59 |
| LDB2 | -0.14 | -0.46 |
| EFNA5 | -0.14 | -0.51 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003723 | RNA binding | IEA | - | |
| GO:0003729 | mRNA binding | TAS | 7692601 | |
| GO:0005515 | protein binding | IPI | 10556305 |16407062 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007417 | central nervous system development | IEA | Brain (GO term level: 6) | - |
| GO:0006810 | transport | IEA | - | |
| GO:0051028 | mRNA transport | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005625 | soluble fraction | TAS | 8401578 | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005654 | nucleoplasm | TAS | 10888599 | |
| GO:0005730 | nucleolus | TAS | 16407062 | |
| GO:0005737 | cytoplasm | TAS | 16407062 | |
| GO:0042788 | polysomal ribosome | TAS | 12837692 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CYFIP1 | FLJ45151 | P140SRA-1 | SHYC | SRA1 | cytoplasmic FMR1 interacting protein 1 | - | HPRD,BioGRID | 11438699 |
| CYFIP2 | PIR121 | cytoplasmic FMR1 interacting protein 2 | - | HPRD,BioGRID | 11438699 |
| FMR1 | FMRP | FRAXA | MGC87458 | POF | POF1 | fragile X mental retardation 1 | Affinity Capture-Western | BioGRID | 10567518 |
| FXR1 | - | fragile X mental retardation, autosomal homolog 1 | - | HPRD,BioGRID | 7489725 |8668200 |
| FXR2 | FMR1L2 | fragile X mental retardation, autosomal homolog 2 | - | HPRD,BioGRID | 7489725 |8668200 |10567518 |
| MAP1B | DKFZp686E1099 | DKFZp686F1345 | FLJ38954 | FUTSCH | MAP5 | microtubule-associated protein 1B | FMR1 (FMRP) interacts with MAP1B mRNA. This interaction was modeled on a demonstrated interaction between FMR1 from an unspecified species and human MAP1B mRNA. | BIND | 15381419 |
| NCL | C23 | FLJ45706 | nucleolin | Affinity Capture-Western | BioGRID | 10567518 |
| NUFIP1 | NUFIP | bA540M5.1 | nuclear fragile X mental retardation protein interacting protein 1 | - | HPRD,BioGRID | 10556305 |
| NUFIP2 | 182-FIP | 82-FIP | FIP-82 | FLJ10976 | KIAA1321 | MGC117262 | PIG1 | nuclear fragile X mental retardation protein interacting protein 2 | - | HPRD,BioGRID | 12837692 |
| RANBP9 | RANBPM | RAN binding protein 9 | FMR1 (FMRP) interacts with RANBP9 (RanBPM). | BIND | 15381419 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | - | HPRD,BioGRID | 11367701 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DAVICIONI PAX FOXO1 SIGNATURE IN ARMS UP | 59 | 38 | All SZGR 2.0 genes in this pathway |
| DAVICIONI TARGETS OF PAX FOXO1 FUSIONS UP | 255 | 177 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS DN | 431 | 263 | All SZGR 2.0 genes in this pathway |
| LAIHO COLORECTAL CANCER SERRATED DN | 86 | 47 | All SZGR 2.0 genes in this pathway |
| SLEBOS HEAD AND NECK CANCER WITH HPV UP | 84 | 43 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL TGFB1 TARGETS UP | 169 | 127 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS BASAL | 330 | 217 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP | 424 | 268 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| HADDAD B LYMPHOCYTE PROGENITOR | 293 | 193 | All SZGR 2.0 genes in this pathway |
| PAL PRMT5 TARGETS UP | 203 | 135 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL | 254 | 164 | All SZGR 2.0 genes in this pathway |
| SANSOM APC MYC TARGETS | 217 | 138 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN | 409 | 268 | All SZGR 2.0 genes in this pathway |
| LABBE WNT3A TARGETS DN | 97 | 53 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR DN | 185 | 116 | All SZGR 2.0 genes in this pathway |
| PYEON CANCER HEAD AND NECK VS CERVICAL UP | 193 | 95 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-101 | 1620 | 1626 | 1A | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-124/506 | 544 | 550 | m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-125/351 | 595 | 601 | 1A | hsa-miR-125bbrain | UCCCUGAGACCCUAACUUGUGA |
| hsa-miR-125abrain | UCCCUGAGACCCUUUAACCUGUG | ||||
| miR-129-5p | 523 | 530 | 1A,m8 | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-130/301 | 2009 | 2015 | 1A | hsa-miR-130abrain | CAGUGCAAUGUUAAAAGGGCAU |
| hsa-miR-301 | CAGUGCAAUAGUAUUGUCAAAGC | ||||
| hsa-miR-130bbrain | CAGUGCAAUGAUGAAAGGGCAU | ||||
| hsa-miR-454-3p | UAGUGCAAUAUUGCUUAUAGGGUUU | ||||
| hsa-miR-130abrain | CAGUGCAAUGUUAAAAGGGCAU | ||||
| hsa-miR-301 | CAGUGCAAUAGUAUUGUCAAAGC | ||||
| hsa-miR-130bbrain | CAGUGCAAUGAUGAAAGGGCAU | ||||
| hsa-miR-454-3p | UAGUGCAAUAUUGCUUAUAGGGUUU | ||||
| miR-132/212 | 1885 | 1891 | 1A | hsa-miR-212SZ | UAACAGUCUCCAGUCACGGCC |
| hsa-miR-132brain | UAACAGUCUACAGCCAUGGUCG | ||||
| miR-136 | 2113 | 2119 | m8 | hsa-miR-136 | ACUCCAUUUGUUUUGAUGAUGGA |
| miR-139 | 1621 | 1628 | 1A,m8 | hsa-miR-139brain | UCUACAGUGCACGUGUCU |
| miR-141/200a | 776 | 782 | 1A | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-144 | 1619 | 1626 | 1A,m8 | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-148/152 | 756 | 763 | 1A,m8 | hsa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| miR-153 | 1753 | 1759 | 1A | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-181 | 1550 | 1556 | 1A | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-182 | 1510 | 1517 | 1A,m8 | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-19 | 1584 | 1590 | 1A | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA | ||||
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-194 | 1886 | 1892 | m8 | hsa-miR-194 | UGUAACAGCAACUCCAUGUGGA |
| hsa-miR-194 | UGUAACAGCAACUCCAUGUGGA | ||||
| miR-200bc/429 | 468 | 474 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-203.1 | 426 | 432 | 1A | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-205 | 1276 | 1282 | 1A | hsa-miR-205 | UCCUUCAUUCCACCGGAGUCUG |
| miR-221/222 | 631 | 638 | 1A,m8 | hsa-miR-221brain | AGCUACAUUGUCUGCUGGGUUUC |
| hsa-miR-222brain | AGCUACAUCUGGCUACUGGGUCUC | ||||
| miR-23 | 1273 | 1279 | 1A | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC | ||||
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-24* | 88 | 95 | 1A,m8 | hsa-miR-189 | GUGCCUACUGAGCUGAUAUCAGU |
| miR-25/32/92/363/367 | 1738 | 1745 | 1A,m8 | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-323 | 2064 | 2070 | 1A | hsa-miR-323brain | GCACAUUACACGGUCGACCUCU |
| miR-324-3p | 273 | 279 | 1A | hsa-miR-324-3p | CCACUGCCCCAGGUGCUGCUGG |
| miR-324-5p | 584 | 590 | 1A | hsa-miR-324-5p | CGCAUCCCCUAGGGCAUUGGUGU |
| miR-335 | 1665 | 1671 | m8 | hsa-miR-335brain | UCAAGAGCAAUAACGAAAAAUGU |
| miR-369-3p | 574 | 580 | 1A | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-374 | 574 | 580 | m8 | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-377 | 493 | 499 | m8 | hsa-miR-377 | AUCACACAAAGGCAACUUUUGU |
| miR-410 | 1298 | 1304 | m8 | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| hsa-miR-410 | AAUAUAACACAGAUGGCCUGU | ||||
| miR-448 | 1753 | 1759 | 1A | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU | ||||
| miR-450 | 523 | 529 | 1A | hsa-miR-450 | UUUUUGCGAUGUGUUCCUAAUA |
| miR-485-3p | 534 | 540 | m8 | hsa-miR-485-3p | GUCAUACACGGCUCUCCUCUCU |
| miR-490 | 1090 | 1096 | 1A | hsa-miR-490 | CAACCUGGAGGACUCCAUGCUG |
| miR-496 | 2237 | 2243 | 1A | hsa-miR-496 | AUUACAUGGCCAAUCUC |
| miR-543 | 621 | 627 | 1A | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
| miR-9 | 75 | 81 | 1A | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
| miR-96 | 1511 | 1517 | 1A | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.