Gene Page: DOCK9
Summary ?
| GeneID | 23348 |
| Symbol | DOCK9 |
| Synonyms | ZIZ1|ZIZIMIN1 |
| Description | dedicator of cytokinesis 9 |
| Reference | MIM:607325|HGNC:HGNC:14132|Ensembl:ENSG00000088387|HPRD:09542|Vega:OTTHUMG00000017260 |
| Gene type | protein-coding |
| Map location | 13q32.3 |
| Pascal p-value | 0.915 |
| Sherlock p-value | 0.112 |
| DMG | 2 (# studies) |
| eGene | Anterior cingulate cortex BA24 Myers' cis & trans |
| Support | CompositeSet Darnell FMRP targets Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 3 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 3 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg21712331 | 13 | 99630489 | DOCK9 | 4.84E-4 | -0.722 | 0.046 | DMG:Wockner_2014 |
| cg10811485 | 13 | 99630583 | DOCK9 | 4.921E-4 | -0.673 | 0.047 | DMG:Wockner_2014 |
| cg04836038 | 13 | 99739382 | DOCK9 | -0.041 | 0.75 | DMG:Nishioka_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16894557 | chr6 | 28999825 | DOCK9 | 23348 | 0.06 | trans | ||
| rs3825428 | 13 | 99398883 | DOCK9 | ENSG00000088387.13 | 1.45694E-6 | 0.04 | 339996 | gtex_brain_ba24 |
| rs3782997 | 13 | 99398932 | DOCK9 | ENSG00000088387.13 | 1.45694E-6 | 0.04 | 339947 | gtex_brain_ba24 |
| rs3782994 | 13 | 99401848 | DOCK9 | ENSG00000088387.13 | 1.45694E-6 | 0.04 | 337031 | gtex_brain_ba24 |
Section II. Transcriptome annotation
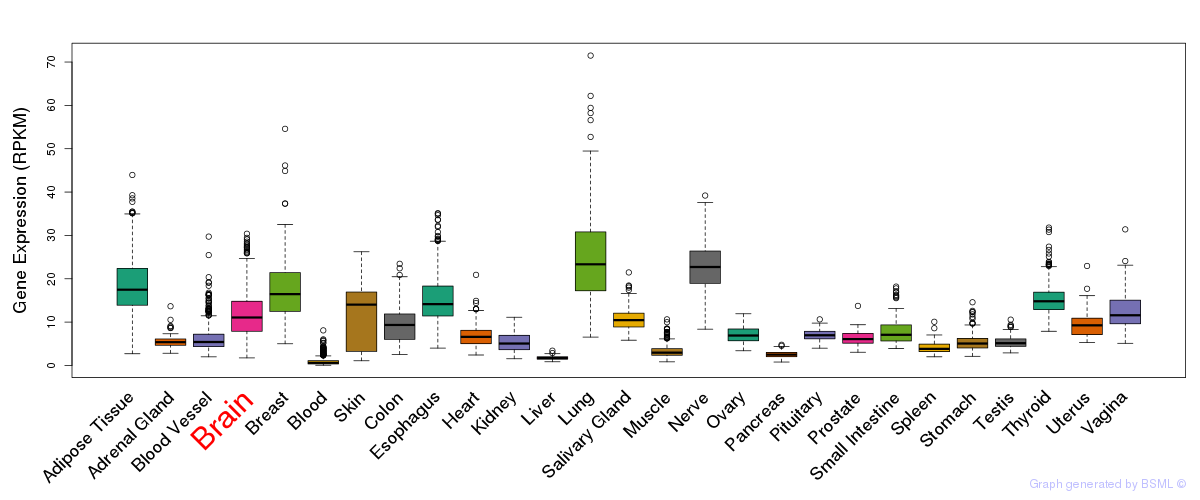
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C11orf82 | 0.87 | 0.83 |
| KNTC1 | 0.87 | 0.81 |
| PCNA | 0.86 | 0.76 |
| FANCD2 | 0.86 | 0.82 |
| C21orf45 | 0.86 | 0.81 |
| PLK4 | 0.85 | 0.83 |
| NCAPG2 | 0.85 | 0.82 |
| GINS2 | 0.85 | 0.60 |
| TIMELESS | 0.85 | 0.74 |
| STIL | 0.84 | 0.81 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.27 | -0.63 | -0.79 |
| AF347015.33 | -0.62 | -0.78 |
| MT-CO2 | -0.62 | -0.79 |
| AF347015.31 | -0.62 | -0.77 |
| MT-CYB | -0.61 | -0.78 |
| AF347015.8 | -0.61 | -0.80 |
| AF347015.15 | -0.61 | -0.79 |
| HLA-F | -0.59 | -0.69 |
| AIFM3 | -0.59 | -0.69 |
| AF347015.2 | -0.59 | -0.77 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005085 | guanyl-nucleotide exchange factor activity | IEA | - | |
| GO:0005525 | GTP binding | IEA | - | |
| GO:0051020 | GTPase binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008150 | biological_process | ND | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0012505 | endomembrane system | IEA | - | |
| GO:0005575 | cellular_component | ND | - | |
| GO:0016020 | membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID CDC42 REG PATHWAY | 30 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | 132 | 101 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS UP | 332 | 228 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 1 UP | 276 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 UP | 341 | 197 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 6HR DN | 167 | 100 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL DN | 226 | 132 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 XPCS DN | 88 | 71 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 TTD DN | 84 | 63 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| GOLDRATH IMMUNE MEMORY | 65 | 42 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 10 | 30 | 17 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL LONG TERM | 302 | 191 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS UP | 175 | 116 | All SZGR 2.0 genes in this pathway |
| CHEN LVAD SUPPORT OF FAILING HEART UP | 103 | 69 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK DN | 318 | 220 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G6 | 153 | 112 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS UP | 317 | 208 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL DN | 216 | 143 | All SZGR 2.0 genes in this pathway |
| KESHELAVA MULTIPLE DRUG RESISTANCE | 88 | 56 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA MF DN | 41 | 28 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS UP | 387 | 225 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 DN | 336 | 211 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION DN | 321 | 200 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| RATTENBACHER BOUND BY CELF1 | 467 | 251 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-136 | 977 | 983 | 1A | hsa-miR-136 | ACUCCAUUUGUUUUGAUGAUGGA |
| miR-148/152 | 472 | 479 | 1A,m8 | hsa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| miR-153 | 929 | 935 | 1A | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-182 | 1039 | 1046 | 1A,m8 | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-203.1 | 854 | 860 | m8 | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-208 | 347 | 353 | 1A | hsa-miR-208 | AUAAGACGAGCAAAAAGCUUGU |
| miR-214 | 1204 | 1210 | m8 | hsa-miR-214brain | ACAGCAGGCACAGACAGGCAG |
| miR-25/32/92/363/367 | 970 | 977 | 1A,m8 | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-409-3p | 554 | 560 | 1A | hsa-miR-409-3p | CGAAUGUUGCUCGGUGAACCCCU |
| miR-410 | 1118 | 1124 | m8 | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| miR-448 | 928 | 935 | 1A,m8 | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-452 | 164 | 170 | m8 | hsa-miR-452 | UGUUUGCAGAGGAAACUGAGAC |
| miR-455 | 551 | 557 | m8 | hsa-miR-455 | UAUGUGCCUUUGGACUACAUCG |
| hsa-miR-455 | UAUGUGCCUUUGGACUACAUCG | ||||
| miR-493-5p | 803 | 809 | m8 | hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU |
| miR-499 | 347 | 353 | 1A | hsa-miR-499 | UUAAGACUUGCAGUGAUGUUUAA |
| miR-9 | 739 | 745 | m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
| miR-96 | 1040 | 1046 | 1A | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.