Gene Page: FN1
Summary ?
| GeneID | 2335 |
| Symbol | FN1 |
| Synonyms | CIG|ED-B|FINC|FN|FNZ|GFND|GFND2|LETS|MSF |
| Description | fibronectin 1 |
| Reference | MIM:135600|HGNC:HGNC:3778|Ensembl:ENSG00000115414|HPRD:00626|Vega:OTTHUMG00000133054 |
| Gene type | protein-coding |
| Map location | 2q34 |
| Pascal p-value | 0.558 |
| Sherlock p-value | 0.423 |
| Fetal beta | 1.153 |
| eGene | Myers' cis & trans |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| DNM:Guipponi_2014 | Whole Exome Sequencing analysis | 49 DNMs were identified by comparing the exome of 53 individuals with sporadic SCZ and of their non-affected parents | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01016 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00916 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| FN1 | chr2 | 216262472 | C | T | NM_002026 NM_212474 NM_212476 NM_212478 NM_212482 | p.1150V>I p.1150V>I p.1150V>I p.1150V>I p.1150V>I | missense missense missense missense missense | Schizophrenia | DNM:Fromer_2014 | ||
| FN1 | G | . | NM_002026 | p.A93LfsX26 | frameshift | NA | NA | Schizophrenia | DNM:Guipponi_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4673840 | chr2 | 215363240 | FN1 | 2335 | 0.18 | cis | ||
| rs4236095 | chr6 | 47368686 | FN1 | 2335 | 0.1 | trans | ||
| rs11002520 | chr10 | 80117056 | FN1 | 2335 | 0.2 | trans | ||
| rs11002521 | chr10 | 80118300 | FN1 | 2335 | 0.09 | trans | ||
| rs1159612 | chr10 | 85329057 | FN1 | 2335 | 0 | trans | ||
| rs11820030 | chr11 | 76133564 | FN1 | 2335 | 0.12 | trans | ||
| rs6095741 | chr20 | 48666589 | FN1 | 2335 | 0.15 | trans | ||
| rs2838712 | chr21 | 46277775 | FN1 | 2335 | 0.14 | trans |
Section II. Transcriptome annotation
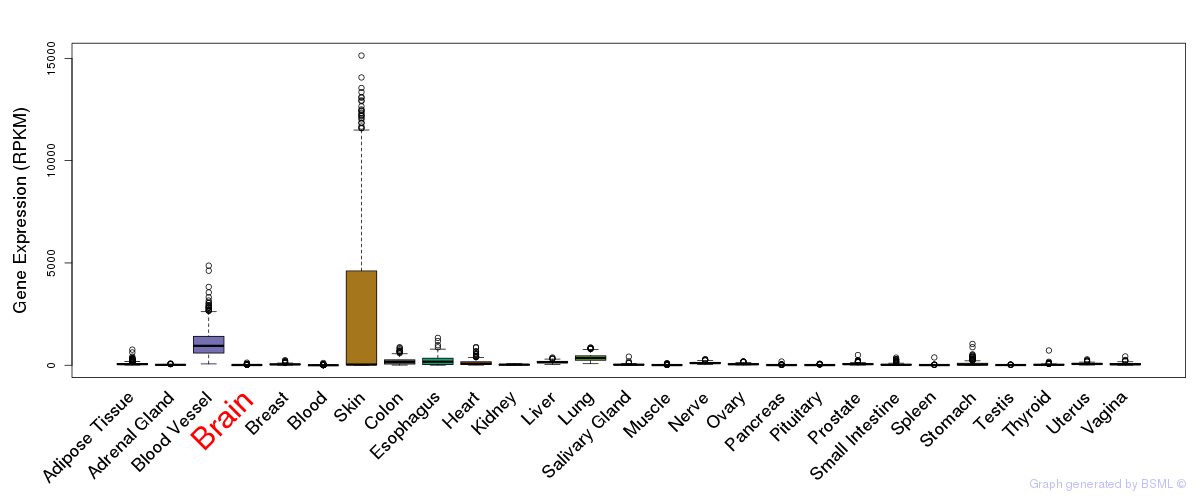
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005518 | collagen binding | NAS | 3024962 | |
| GO:0005201 | extracellular matrix structural constituent | NAS | - | |
| GO:0008201 | heparin binding | NAS | 10075919 | |
| GO:0016491 | oxidoreductase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | NAS | 15609325 | |
| GO:0007155 | cell adhesion | NAS | 1423622 | |
| GO:0009611 | response to wounding | NAS | 7989369 | |
| GO:0008152 | metabolic process | IEA | - | |
| GO:0006953 | acute-phase response | IEA | - | |
| GO:0016477 | cell migration | NAS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005793 | ER-Golgi intermediate compartment | IDA | 15308636 | |
| GO:0005576 | extracellular region | EXP | 444675 | |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005576 | extracellular region | NAS | 14718574 | |
| GO:0005578 | proteinaceous extracellular matrix | NAS | - | |
| GO:0031093 | platelet alpha granule lumen | EXP | 444675 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AMBP | EDC1 | HCP | HI30 | IATIL | ITI | ITIL | ITILC | UTI | alpha-1-microglobulin/bikunin precursor | - | HPRD,BioGRID | 7519849 |
| APCS | MGC88159 | PTX2 | SAP | amyloid P component, serum | - | HPRD,BioGRID | 12100475 |
| C1QA | - | complement component 1, q subcomponent, A chain | - | HPRD,BioGRID | 6981115 |
| CD44 | CDW44 | CSPG8 | ECMR-III | HCELL | IN | LHR | MC56 | MDU2 | MDU3 | MGC10468 | MIC4 | MUTCH-I | Pgp1 | CD44 molecule (Indian blood group) | Affinity Capture-Western Reconstituted Complex | BioGRID | 1730778 |
| COL13A1 | COLXIIIA1 | FLJ42485 | collagen, type XIII, alpha 1 | - | HPRD,BioGRID | 11956183 |
| COL1A1 | OI4 | collagen, type I, alpha 1 | - | HPRD,BioGRID | 8468356 |
| COL1A2 | OI4 | collagen, type I, alpha 2 | - | HPRD,BioGRID | 8468356 |
| COL2A1 | ANFH | AOM | COL11A3 | MGC131516 | SEDC | collagen, type II, alpha 1 | - | HPRD,BioGRID | 2229073 |3997552 |
| COL4A1 | arresten | collagen, type IV, alpha 1 | - | HPRD | 3997552 |
| COL4A2 | DKFZp686I14213 | FLJ22259 | collagen, type IV, alpha 2 | - | HPRD | 3997552 |
| COL4A3 | - | collagen, type IV, alpha 3 (Goodpasture antigen) | - | HPRD | 3997552 |
| COL4A4 | CA44 | collagen, type IV, alpha 4 | - | HPRD | 3997552 |
| COL4A5 | ASLN | ATS | CA54 | MGC167109 | MGC42377 | collagen, type IV, alpha 5 | - | HPRD | 3997552 |
| COL4A6 | MGC88184 | collagen, type IV, alpha 6 | - | HPRD | 3997552 |
| COL7A1 | EBD1 | EBDCT | EBR1 | collagen, type VII, alpha 1 | - | HPRD,BioGRID | 7963647 |9169408 |
| CRP | MGC149895 | MGC88244 | PTX1 | C-reactive protein, pentraxin-related | - | HPRD,BioGRID | 6693419 |
| CTSD | CLN10 | CPSD | MGC2311 | cathepsin D | - | HPRD | 3988746 |
| CXCL12 | PBSF | SCYB12 | SDF-1a | SDF-1b | SDF1 | SDF1A | SDF1B | TLSF-a | TLSF-b | TPAR1 | chemokine (C-X-C motif) ligand 12 (stromal cell-derived factor 1) | - | HPRD,BioGRID | 11023498 |
| DCN | CSCD | DSPG2 | PG40 | PGII | PGS2 | SLRR1B | decorin | - | HPRD | 1468447 |
| F13A1 | F13A | coagulation factor XIII, A1 polypeptide | - | HPRD,BioGRID | 8905624 |
| FASLG | APT1LG1 | CD178 | CD95L | FASL | TNFSF6 | Fas ligand (TNF superfamily, member 6) | - | HPRD,BioGRID | 11276204 |
| FBLN1 | FBLN | fibulin 1 | - | HPRD,BioGRID | 1400330 |
| FBLN2 | - | fibulin 2 | - | HPRD | 10848816 |
| GSN | DKFZp313L0718 | gelsolin (amyloidosis, Finnish type) | - | HPRD,BioGRID | 6092370 |
| HSPG2 | PLC | PRCAN | SJA | SJS | SJS1 | heparan sulfate proteoglycan 2 | Reconstituted Complex | BioGRID | 11493006 |
| IGFBP3 | BP-53 | IBP3 | insulin-like growth factor binding protein 3 | - | HPRD,BioGRID | 11344214 |
| ITGA3 | CD49C | FLJ34631 | FLJ34704 | GAP-B3 | GAPB3 | MSK18 | VCA-2 | VL3A | VLA3a | integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) | - | HPRD | 9733622 |
| ITGA4 | CD49D | IA4 | MGC90518 | integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) | - | HPRD | 9117345 |11010812 |
| ITGA5 | CD49e | FNRA | VLA5A | integrin, alpha 5 (fibronectin receptor, alpha polypeptide) | Reconstituted Complex | BioGRID | 1532572 |
| ITGA5 | CD49e | FNRA | VLA5A | integrin, alpha 5 (fibronectin receptor, alpha polypeptide) | - | HPRD | 9211865 |11801679 |
| ITGA8 | - | integrin, alpha 8 | - | HPRD | 7559467 |
| ITGB6 | - | integrin, beta 6 | - | HPRD,BioGRID | 1532572 |8120056 |
| ITGB7 | - | integrin, beta 7 | - | HPRD,BioGRID | 1372909 |
| KLK3 | APS | KLK2A1 | PSA | hK3 | kallikrein-related peptidase 3 | - | HPRD | 9016396 |
| LACRT | MGC71934 | lacritin | - | HPRD,BioGRID | 11419941 |
| LGALS3BP | 90K | BTBD17B | MAC-2-BP | lectin, galactoside-binding, soluble, 3 binding protein | - | HPRD | 9501082 |
| LPA | AK38 | APOA | LP | lipoprotein, Lp(a) | - | HPRD,BioGRID | 2531657 |
| LRG1 | HMFT1766 | LRG | leucine-rich alpha-2-glycoprotein 1 | - | HPRD | 11801638 |
| LTBP1 | MGC163161 | latent transforming growth factor beta binding protein 1 | - | HPRD | 8756760 |
| MAG | GMA | S-MAG | SIGLEC-4A | SIGLEC4A | myelin associated glycoprotein | - | HPRD | 11423128 |
| MATN2 | - | matrilin 2 | - | HPRD | 12180907 |
| MEP1A | PPHA | meprin A, alpha (PABA peptide hydrolase) | - | HPRD,BioGRID | 8063866 |
| MEP1B | - | meprin A, beta | - | HPRD,BioGRID | 8063866 |
| MIA | CD-RAP | melanoma inhibitory activity | - | HPRD,BioGRID | 11157741 |
| MMP9 | CLG4B | GELB | MMP-9 | matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) | - | HPRD | 11134254 |
| MYOC | GLC1A | GPOA | JOAG | JOAG1 | TIGR | myocilin | myocilin, trabecular meshwork inducible glucocorticoid response | - | HPRD,BioGRID | 11773026 |
| NOV | CCN3 | IGFBP9 | nephroblastoma overexpressed gene | - | HPRD,BioGRID | 12902636 |
| PKD1 | PBP | polycystic kidney disease 1 (autosomal dominant) | - | HPRD,BioGRID | 11752017 |
| PRELP | MGC45323 | MST161 | MSTP161 | SLRR2A | proline/arginine-rich end leucine-rich repeat protein | - | HPRD | 11847210 |
| REG3A | HIP | INGAP | PAP | PAP-H | PAP1 | PBCGF | REG-III | REG3 | regenerating islet-derived 3 alpha | - | HPRD,BioGRID | 8997243 |
| SCGB1A1 | CC10 | CC16 | CCSP | UGB | secretoglobin, family 1A, member 1 (uteroglobin) | - | HPRD | 10470078 |
| SDC2 | HSPG | HSPG1 | SYND2 | syndecan 2 | - | HPRD,BioGRID | 10772816 |
| SFRP2 | FRP-2 | SARP1 | SDF-5 | secreted frizzled-related protein 2 | SFRP2 interacts with an unspecified isoform of FN1 (FN). This interaction was modeled on a demonstrated interaction between canine SFRP2 and human FN1. | BIND | 14709558 |
| TAC1 | Hs.2563 | NK2 | NKNA | NPK | TAC2 | tachykinin, precursor 1 | - | HPRD | 11342427 |
| TGFBI | BIGH3 | CDB1 | CDG2 | CDGG1 | CSD | CSD1 | CSD2 | CSD3 | EBMD | LCD1 | transforming growth factor, beta-induced, 68kDa | - | HPRD,BioGRID | 11867580 |12034705 |
| TGM2 | G-ALPHA-h | GNAH | TG2 | TGC | transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase) | - | HPRD,BioGRID | 11686302 |11829448 |
| THBS1 | THBS | TSP | TSP1 | thrombospondin 1 | - | HPRD | 11773026 |
| TMPRSS6 | IRIDA | transmembrane protease, serine 6 | - | HPRD,BioGRID | 12149247 |
| TNC | HXB | MGC167029 | TN | tenascin C | - | HPRD,BioGRID | 7499434 |
| TSHR | CHNG1 | LGR3 | MGC75129 | hTSHR-I | thyroid stimulating hormone receptor | - | HPRD,BioGRID | 11981027 |
| VHL | HRCA1 | RCA1 | VHL1 | von Hippel-Lindau tumor suppressor | pVHL interacts with fibronectin 1 | BIND | 10823831 |
| VHL | HRCA1 | RCA1 | VHL1 | von Hippel-Lindau tumor suppressor | - | HPRD,BioGRID | 9651579 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG FOCAL ADHESION | 201 | 138 | All SZGR 2.0 genes in this pathway |
| KEGG ECM RECEPTOR INTERACTION | 84 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG SMALL CELL LUNG CANCER | 84 | 67 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN1 PATHWAY | 66 | 44 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN3 PATHWAY | 43 | 25 | All SZGR 2.0 genes in this pathway |
| PID ANGIOPOIETIN RECEPTOR PATHWAY | 50 | 41 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN A9B1 PATHWAY | 25 | 18 | All SZGR 2.0 genes in this pathway |
| PID AVB3 INTEGRIN PATHWAY | 75 | 53 | All SZGR 2.0 genes in this pathway |
| PID SYNDECAN 4 PATHWAY | 32 | 25 | All SZGR 2.0 genes in this pathway |
| PID UPA UPAR PATHWAY | 42 | 30 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN5 PATHWAY | 17 | 9 | All SZGR 2.0 genes in this pathway |
| PID SYNDECAN 2 PATHWAY | 33 | 27 | All SZGR 2.0 genes in this pathway |
| PID LYMPH ANGIOGENESIS PATHWAY | 25 | 16 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN A4B1 PATHWAY | 33 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | 91 | 65 | All SZGR 2.0 genes in this pathway |
| REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | 89 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRIN CELL SURFACE INTERACTIONS | 79 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | 15 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | 15 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | 27 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET AGGREGATION PLUG FORMATION | 36 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL UP | 285 | 181 | All SZGR 2.0 genes in this pathway |
| PICCALUGA ANGIOIMMUNOBLASTIC LYMPHOMA UP | 205 | 140 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE UP | 351 | 230 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA UP | 294 | 178 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST DUCTAL CARCINOMA VS DUCTAL NORMAL UP | 44 | 25 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST DUCTAL CARCINOMA VS LOBULAR NORMAL UP | 73 | 50 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST LOBULAR CARCINOMA VS DUCTAL NORMAL UP | 69 | 38 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST LOBULAR CARCINOMA VS LOBULAR NORMAL DN | 74 | 42 | All SZGR 2.0 genes in this pathway |
| WILCOX RESPONSE TO PROGESTERONE DN | 66 | 44 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| SAMOLS TARGETS OF KHSV MIRNAS DN | 62 | 35 | All SZGR 2.0 genes in this pathway |
| HOOI ST7 TARGETS DN | 123 | 78 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| RODRIGUES NTN1 TARGETS DN | 158 | 102 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER ADVANCED VS EARLY UP | 175 | 120 | All SZGR 2.0 genes in this pathway |
| JAEGER METASTASIS UP | 44 | 23 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 10D UP | 194 | 122 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 16D UP | 175 | 108 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP | 457 | 269 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS UP | 238 | 144 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION DN | 329 | 219 | All SZGR 2.0 genes in this pathway |
| SABATES COLORECTAL ADENOMA DN | 291 | 176 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA DN | 77 | 52 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| VANHARANTA UTERINE FIBROID UP | 45 | 26 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| WANG ESOPHAGUS CANCER VS NORMAL UP | 121 | 72 | All SZGR 2.0 genes in this pathway |
| JOHANSSON GLIOMAGENESIS BY PDGFB UP | 58 | 44 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 3 4WK UP | 214 | 144 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK UP | 271 | 175 | All SZGR 2.0 genes in this pathway |
| VETTER TARGETS OF PRKCA AND ETS1 UP | 16 | 8 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL TGFB1 TARGETS UP | 169 | 127 | All SZGR 2.0 genes in this pathway |
| BEGUM TARGETS OF PAX3 FOXO1 FUSION AND PAX3 | 7 | 5 | All SZGR 2.0 genes in this pathway |
| TSUNODA CISPLATIN RESISTANCE DN | 51 | 38 | All SZGR 2.0 genes in this pathway |
| WANG HCP PROSTATE CANCER | 111 | 69 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| SIMBULAN UV RESPONSE IMMORTALIZED DN | 31 | 26 | All SZGR 2.0 genes in this pathway |
| KIM MYCN AMPLIFICATION TARGETS DN | 103 | 59 | All SZGR 2.0 genes in this pathway |
| TSAI RESPONSE TO IONIZING RADIATION | 149 | 101 | All SZGR 2.0 genes in this pathway |
| KARAKAS TGFB1 SIGNALING | 18 | 15 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| GUENTHER GROWTH SPHERICAL VS ADHERENT DN | 26 | 19 | All SZGR 2.0 genes in this pathway |
| GOUYER TATI TARGETS DN | 17 | 12 | All SZGR 2.0 genes in this pathway |
| FRIDMAN SENESCENCE UP | 77 | 60 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN | 411 | 249 | All SZGR 2.0 genes in this pathway |
| WU CELL MIGRATION | 184 | 114 | All SZGR 2.0 genes in this pathway |
| COLIN PILOCYTIC ASTROCYTOMA VS GLIOBLASTOMA DN | 28 | 21 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION UP | 69 | 55 | All SZGR 2.0 genes in this pathway |
| PETRETTO CARDIAC HYPERTROPHY | 34 | 26 | All SZGR 2.0 genes in this pathway |
| AMIT SERUM RESPONSE 60 MCF10A | 57 | 42 | All SZGR 2.0 genes in this pathway |
| WANG PROSTATE CANCER ANDROGEN INDEPENDENT | 66 | 37 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 UP | 256 | 159 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS SUBSET | 33 | 20 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS UP | 266 | 171 | All SZGR 2.0 genes in this pathway |
| NELSON RESPONSE TO ANDROGEN DN | 19 | 14 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT UP | 197 | 110 | All SZGR 2.0 genes in this pathway |
| IIZUKA LIVER CANCER PROGRESSION L0 L1 UP | 17 | 12 | All SZGR 2.0 genes in this pathway |
| CHANG POU5F1 TARGETS UP | 15 | 10 | All SZGR 2.0 genes in this pathway |
| KENNY CTNNB1 TARGETS DN | 52 | 34 | All SZGR 2.0 genes in this pathway |
| GILDEA METASTASIS | 30 | 14 | All SZGR 2.0 genes in this pathway |
| LEI MYB TARGETS | 318 | 215 | All SZGR 2.0 genes in this pathway |
| YAO HOXA10 TARGETS VIA PROGESTERONE DN | 19 | 12 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| MODY HIPPOCAMPUS NEONATAL | 35 | 25 | All SZGR 2.0 genes in this pathway |
| JI RESPONSE TO FSH DN | 58 | 43 | All SZGR 2.0 genes in this pathway |
| LINDVALL IMMORTALIZED BY TERT DN | 80 | 56 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA RESPONSE TO TGFB1 C2 | 25 | 18 | All SZGR 2.0 genes in this pathway |
| GERHOLD ADIPOGENESIS DN | 64 | 44 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA EARLY RESPONSE TO TGFB1 | 58 | 43 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY NO BLOOD UP | 222 | 139 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| KAYO CALORIE RESTRICTION MUSCLE UP | 95 | 64 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA CANCER | 83 | 52 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS UP | 280 | 183 | All SZGR 2.0 genes in this pathway |
| URS ADIPOCYTE DIFFERENTIATION DN | 30 | 20 | All SZGR 2.0 genes in this pathway |
| BILD HRAS ONCOGENIC SIGNATURE | 261 | 166 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND MACROPHAGE | 77 | 50 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND ENDOTHELIUM | 66 | 47 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND FIBROBLAST | 132 | 93 | All SZGR 2.0 genes in this pathway |
| JI CARCINOGENESIS BY KRAS AND STK11 DN | 17 | 12 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE DN | 195 | 135 | All SZGR 2.0 genes in this pathway |
| KYNG ENVIRONMENTAL STRESS RESPONSE UP | 56 | 41 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS UP | 317 | 208 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| DE YY1 TARGETS DN | 92 | 64 | All SZGR 2.0 genes in this pathway |
| OUYANG PROSTATE CANCER MARKERS | 19 | 16 | All SZGR 2.0 genes in this pathway |
| ENGELMANN CANCER PROGENITORS DN | 70 | 44 | All SZGR 2.0 genes in this pathway |
| HUANG FOXA2 TARGETS UP | 45 | 28 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| BRUECKNER TARGETS OF MIRLET7A3 DN | 78 | 49 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA METAGENE | 242 | 168 | All SZGR 2.0 genes in this pathway |
| CROMER TUMORIGENESIS UP | 63 | 36 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| GU PDEF TARGETS UP | 71 | 49 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| VANASSE BCL2 TARGETS DN | 74 | 50 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS KERATINOCYTE UP | 91 | 63 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS FIBROBLAST UP | 84 | 60 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| LIU VAV3 PROSTATE CARCINOGENESIS UP | 89 | 61 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE UP | 181 | 106 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER UP | 288 | 168 | All SZGR 2.0 genes in this pathway |
| SCHOEN NFKB SIGNALING | 34 | 26 | All SZGR 2.0 genes in this pathway |
| KESHELAVA MULTIPLE DRUG RESISTANCE | 88 | 56 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE 2 | 277 | 172 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT DN | 222 | 141 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN | 253 | 192 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS UP | 221 | 135 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| QI HYPOXIA TARGETS OF HIF1A AND FOXA2 | 37 | 27 | All SZGR 2.0 genes in this pathway |
| HOELZEL NF1 TARGETS DN | 115 | 73 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN | 505 | 328 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| ZHU SKIL TARGETS UP | 20 | 14 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS DN | 315 | 215 | All SZGR 2.0 genes in this pathway |
| AZARE NEOPLASTIC TRANSFORMATION BY STAT3 UP | 121 | 70 | All SZGR 2.0 genes in this pathway |
| AZARE STAT3 TARGETS | 24 | 12 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 4 | 110 | 66 | All SZGR 2.0 genes in this pathway |
| DELACROIX RARG BOUND MEF | 367 | 231 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR BOUND ES | 462 | 273 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 | 227 | 149 | All SZGR 2.0 genes in this pathway |
| ANASTASSIOU CANCER MESENCHYMAL TRANSITION SIGNATURE | 64 | 40 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
| NABA ECM GLYCOPROTEINS | 196 | 99 | All SZGR 2.0 genes in this pathway |
| NABA CORE MATRISOME | 275 | 148 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-1/206 | 683 | 690 | 1A,m8 | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-200bc/429 | 531 | 537 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-217 | 573 | 579 | m8 | hsa-miR-217 | UACUGCAUCAGGAACUGAUUGGAU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.