Gene Page: HAAO
Summary ?
| GeneID | 23498 |
| Symbol | HAAO |
| Synonyms | 3-HAO|HAO |
| Description | 3-hydroxyanthranilate 3,4-dioxygenase |
| Reference | MIM:604521|HGNC:HGNC:4796|Ensembl:ENSG00000162882|HPRD:05158|Vega:OTTHUMG00000152348 |
| Gene type | protein-coding |
| Map location | 2p21 |
| Pascal p-value | 0.054 |
| Fetal beta | -0.235 |
| eGene | Caudate basal ganglia Hippocampus Hypothalamus Nucleus accumbens basal ganglia Putamen basal ganglia |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6734957 | 2 | 42813247 | HAAO | ENSG00000162882.10 | 2.703E-6 | 0 | 206485 | gtex_brain_putamen_basal |
| rs6715204 | 2 | 43088470 | HAAO | ENSG00000162882.10 | 2.978E-6 | 0 | -68738 | gtex_brain_putamen_basal |
| rs7590408 | 2 | 43096490 | HAAO | ENSG00000162882.10 | 6.94E-7 | 0 | -76758 | gtex_brain_putamen_basal |
| rs7580643 | 2 | 43097928 | HAAO | ENSG00000162882.10 | 8.571E-7 | 0 | -78196 | gtex_brain_putamen_basal |
| rs4597584 | 2 | 43101457 | HAAO | ENSG00000162882.10 | 1.046E-6 | 0 | -81725 | gtex_brain_putamen_basal |
| rs4952946 | 2 | 43106093 | HAAO | ENSG00000162882.10 | 2.333E-8 | 0 | -86361 | gtex_brain_putamen_basal |
| rs4594497 | 2 | 43106277 | HAAO | ENSG00000162882.10 | 2.059E-8 | 0 | -86545 | gtex_brain_putamen_basal |
| rs9917172 | 2 | 43110476 | HAAO | ENSG00000162882.10 | 2.813E-8 | 0 | -90744 | gtex_brain_putamen_basal |
| rs4350800 | 2 | 43113206 | HAAO | ENSG00000162882.10 | 1.469E-7 | 0 | -93474 | gtex_brain_putamen_basal |
| rs4356707 | 2 | 43114168 | HAAO | ENSG00000162882.10 | 2.869E-8 | 0 | -94436 | gtex_brain_putamen_basal |
| rs58585549 | 2 | 43117246 | HAAO | ENSG00000162882.10 | 2.638E-8 | 0 | -97514 | gtex_brain_putamen_basal |
| rs6544610 | 2 | 43120250 | HAAO | ENSG00000162882.10 | 1.988E-6 | 0 | -100518 | gtex_brain_putamen_basal |
| rs4953675 | 2 | 43121284 | HAAO | ENSG00000162882.10 | 6.752E-7 | 0 | -101552 | gtex_brain_putamen_basal |
| rs6544611 | 2 | 43122486 | HAAO | ENSG00000162882.10 | 6.752E-7 | 0 | -102754 | gtex_brain_putamen_basal |
| rs6544612 | 2 | 43122766 | HAAO | ENSG00000162882.10 | 6.752E-7 | 0 | -103034 | gtex_brain_putamen_basal |
| rs11883440 | 2 | 43123189 | HAAO | ENSG00000162882.10 | 6.939E-7 | 0 | -103457 | gtex_brain_putamen_basal |
| rs7569325 | 2 | 43124989 | HAAO | ENSG00000162882.10 | 7.91E-7 | 0 | -105257 | gtex_brain_putamen_basal |
| rs4280501 | 2 | 43127045 | HAAO | ENSG00000162882.10 | 2.527E-6 | 0 | -107313 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
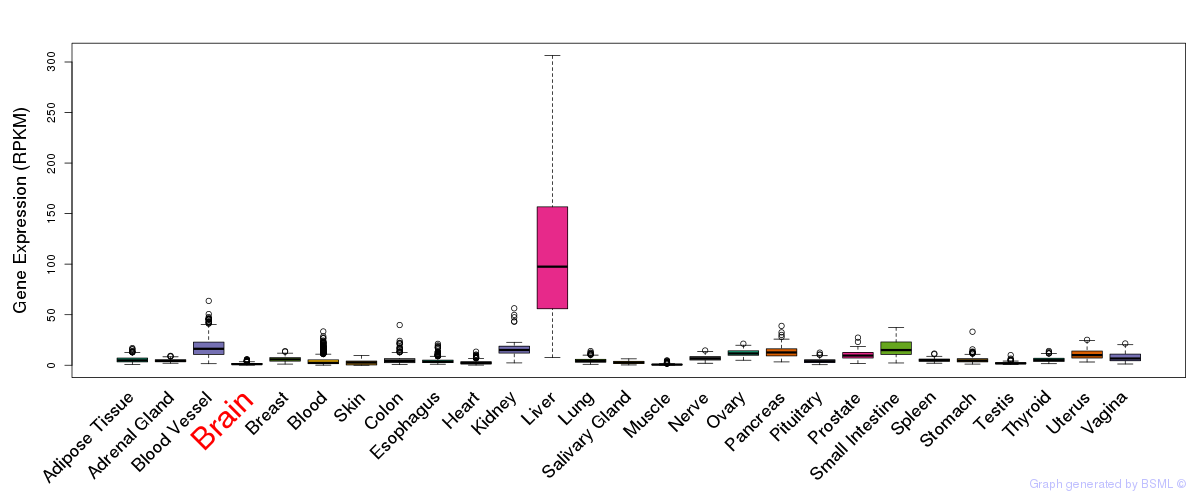
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG TRYPTOPHAN METABOLISM | 40 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | 200 | 136 | All SZGR 2.0 genes in this pathway |
| REACTOME TRYPTOPHAN CATABOLISM | 11 | 9 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP | 390 | 236 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM4 | 261 | 153 | All SZGR 2.0 genes in this pathway |
| ALCALAY AML BY NPM1 LOCALIZATION DN | 184 | 132 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN | 354 | 216 | All SZGR 2.0 genes in this pathway |
| PIONTEK PKD1 TARGETS UP | 38 | 24 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS PROLIFERATION DN | 179 | 97 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS POLYSOMY7 UP | 79 | 50 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER LATE RECURRENCE DN | 69 | 48 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SURVIVAL DN | 113 | 76 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S3 | 266 | 180 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA DN | 267 | 160 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT DN | 222 | 141 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP | 307 | 182 | All SZGR 2.0 genes in this pathway |
| OHGUCHI LIVER HNF4A TARGETS DN | 149 | 85 | All SZGR 2.0 genes in this pathway |