| BIOCARTA RANKL PATHWAY
| 14 | 13 | All SZGR 2.0 genes in this pathway |
| PID FRA PATHWAY
| 37 | 28 | All SZGR 2.0 genes in this pathway |
| PID AP1 PATHWAY
| 70 | 60 | All SZGR 2.0 genes in this pathway |
| PID DELTA NP63 PATHWAY
| 47 | 34 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN
| 634 | 384 | All SZGR 2.0 genes in this pathway |
| PICCALUGA ANGIOIMMUNOBLASTIC LYMPHOMA DN
| 136 | 86 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT EARLY UP
| 21 | 19 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA MYELOID UP
| 30 | 19 | All SZGR 2.0 genes in this pathway |
| LIU CMYB TARGETS UP
| 165 | 106 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN
| 460 | 312 | All SZGR 2.0 genes in this pathway |
| RODRIGUES DCC TARGETS DN
| 121 | 84 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS TURQUOISE UP
| 79 | 50 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS MAGENTA UP
| 28 | 18 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY UP
| 78 | 41 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 UP
| 380 | 236 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 UP
| 418 | 263 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS DN
| 240 | 171 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS UP
| 214 | 155 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN
| 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN
| 770 | 415 | All SZGR 2.0 genes in this pathway |
| TANG SENESCENCE TP53 TARGETS DN
| 57 | 33 | All SZGR 2.0 genes in this pathway |
| WANG ESOPHAGUS CANCER VS NORMAL DN
| 101 | 66 | All SZGR 2.0 genes in this pathway |
| DUNNE TARGETS OF AML1 MTG8 FUSION DN
| 19 | 17 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION
| 77 | 51 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION CCNE1
| 40 | 26 | All SZGR 2.0 genes in this pathway |
| EBAUER TARGETS OF PAX3 FOXO1 FUSION UP
| 207 | 128 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA COAMPLIFIED WITH MYCN
| 43 | 29 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS UP
| 380 | 213 | All SZGR 2.0 genes in this pathway |
| WANG HCP PROSTATE CANCER
| 111 | 69 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP
| 811 | 508 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX
| 795 | 478 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS DN
| 261 | 155 | All SZGR 2.0 genes in this pathway |
| BALDWIN PRKCI TARGETS UP
| 35 | 26 | All SZGR 2.0 genes in this pathway |
| AMIT DELAYED EARLY GENES
| 18 | 15 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 60 HELA
| 46 | 32 | All SZGR 2.0 genes in this pathway |
| KANNAN TP53 TARGETS DN
| 21 | 15 | All SZGR 2.0 genes in this pathway |
| PENG GLUCOSE DEPRIVATION DN
| 169 | 112 | All SZGR 2.0 genes in this pathway |
| MA MYELOID DIFFERENTIATION DN
| 44 | 30 | All SZGR 2.0 genes in this pathway |
| SCHURINGA STAT5A TARGETS DN
| 18 | 10 | All SZGR 2.0 genes in this pathway |
| NATSUME RESPONSE TO INTERFERON BETA UP
| 71 | 49 | All SZGR 2.0 genes in this pathway |
| HENDRICKS SMARCA4 TARGETS UP
| 56 | 35 | All SZGR 2.0 genes in this pathway |
| LEONARD HYPOXIA
| 47 | 35 | All SZGR 2.0 genes in this pathway |
| CHEN LVAD SUPPORT OF FAILING HEART UP
| 103 | 69 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D5
| 39 | 26 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN
| 312 | 203 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 1
| 33 | 24 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA NORMAL
| 311 | 205 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN
| 434 | 302 | All SZGR 2.0 genes in this pathway |
| SASSON FSH RESPONSE
| 9 | 6 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP
| 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP
| 783 | 442 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED
| 1229 | 713 | All SZGR 2.0 genes in this pathway |
| SARRIO EPITHELIAL MESENCHYMAL TRANSITION DN
| 154 | 101 | All SZGR 2.0 genes in this pathway |
| ZHENG GLIOBLASTOMA PLASTICITY DN
| 58 | 39 | All SZGR 2.0 genes in this pathway |
| RAY TUMORIGENESIS BY ERBB2 CDC25A DN
| 159 | 105 | All SZGR 2.0 genes in this pathway |
| CHEN HOXA5 TARGETS 9HR UP
| 223 | 132 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C D DN
| 252 | 155 | All SZGR 2.0 genes in this pathway |
| ZAIDI OSTEOBLAST TRANSCRIPTION FACTORS
| 14 | 12 | All SZGR 2.0 genes in this pathway |
| COULOUARN TEMPORAL TGFB1 SIGNATURE UP
| 109 | 68 | All SZGR 2.0 genes in this pathway |
| UZONYI RESPONSE TO LEUKOTRIENE AND THROMBIN
| 37 | 26 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN
| 253 | 192 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP
| 307 | 182 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION
| 456 | 287 | All SZGR 2.0 genes in this pathway |
| PANGAS TUMOR SUPPRESSION BY SMAD1 AND SMAD5 DN
| 157 | 106 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN
| 918 | 550 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS DN
| 315 | 215 | All SZGR 2.0 genes in this pathway |
| DALESSIO TSA RESPONSE
| 29 | 16 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP
| 682 | 433 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3
| 824 | 528 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D
| 658 | 397 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE VIA P38 COMPLETE
| 227 | 151 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE VIA P38 PARTIAL
| 160 | 106 | All SZGR 2.0 genes in this pathway |
| RATTENBACHER BOUND BY CELF1
| 467 | 251 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL UP
| 289 | 184 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 3 TRANSIENTLY INDUCED BY EGF
| 222 | 159 | All SZGR 2.0 genes in this pathway |
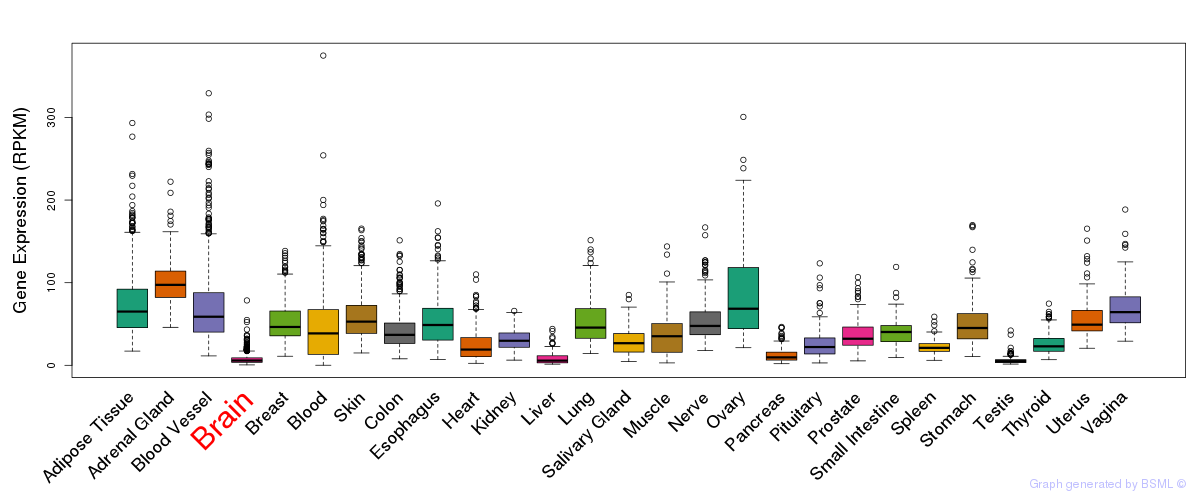
 Differentially methylated gene
Differentially methylated gene eQTL annotation
eQTL annotation