Gene Page: APOL2
Summary ?
| GeneID | 23780 |
| Symbol | APOL2 |
| Synonyms | APOL-II|APOL3 |
| Description | apolipoprotein L2 |
| Reference | MIM:607252|HGNC:HGNC:619|Ensembl:ENSG00000128335|HPRD:06263|Vega:OTTHUMG00000150634 |
| Gene type | protein-coding |
| Map location | 22q12 |
| Pascal p-value | 0.201 |
| Sherlock p-value | 0.953 |
| Fetal beta | -0.122 |
| eGene | Myers' cis & trans Meta |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4820222 | chr22 | 36609738 | APOL2 | 23780 | 4.26E-4 | cis | ||
| rs11748886 | chr5 | 167702040 | APOL2 | 23780 | 0.08 | trans | ||
| rs17235614 | chr13 | 97953192 | APOL2 | 23780 | 0.11 | trans | ||
| rs4820222 | chr22 | 36609738 | APOL2 | 23780 | 0.03 | trans |
Section II. Transcriptome annotation
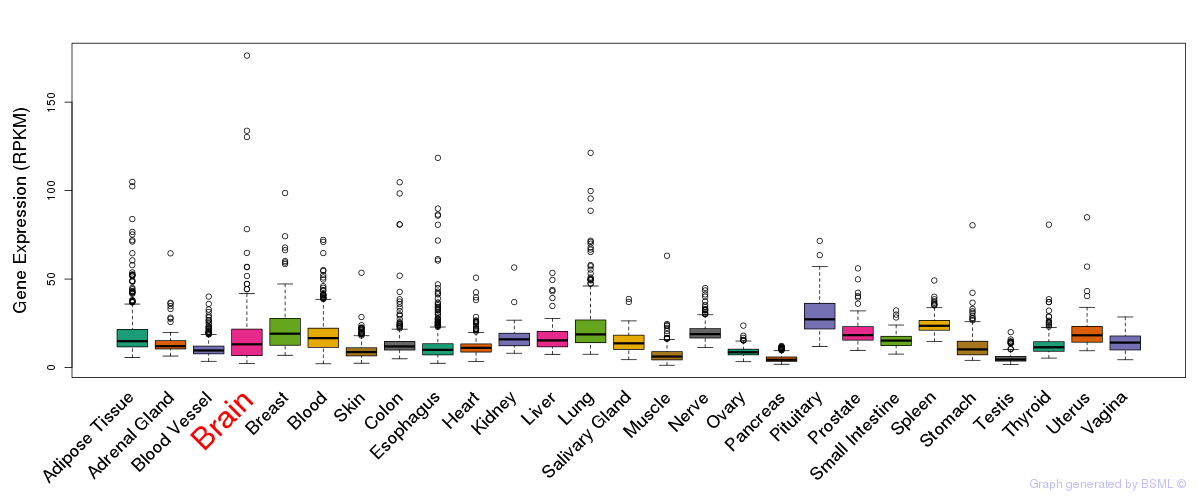
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005102 | receptor binding | NAS | Neurotransmitter (GO term level: 4) | 11374903 |
| GO:0008289 | lipid binding | NAS | 11374903 | |
| GO:0008035 | high-density lipoprotein binding | NAS | 11374903 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008203 | cholesterol metabolic process | NAS | 11374903 | |
| GO:0006953 | acute-phase response | NAS | 11374903 | |
| GO:0006629 | lipid metabolic process | NAS | 11374903 |11944986 | |
| GO:0006869 | lipid transport | NAS | 11374903 | |
| GO:0007275 | multicellular organismal development | NAS | 11374903 | |
| GO:0042157 | lipoprotein metabolic process | IEA | - | |
| GO:0060135 | maternal process involved in pregnancy | NAS | 11374903 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005789 | endoplasmic reticulum membrane | NAS | 11374903 | |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| ROPERO HDAC2 TARGETS | 114 | 71 | All SZGR 2.0 genes in this pathway |
| SANA RESPONSE TO IFNG UP | 78 | 50 | All SZGR 2.0 genes in this pathway |
| OKUMURA INFLAMMATORY RESPONSE LPS | 183 | 115 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K27ME3 DN | 228 | 114 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER DN | 238 | 145 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |