Gene Page: CADM2
Summary ?
| GeneID | 253559 |
| Symbol | CADM2 |
| Synonyms | IGSF4D|NECL3|Necl-3|SynCAM 2|synCAM2 |
| Description | cell adhesion molecule 2 |
| Reference | MIM:609938|HGNC:HGNC:29849|Ensembl:ENSG00000175161|HPRD:11046|Vega:OTTHUMG00000158990 |
| Gene type | protein-coding |
| Map location | 3p12.1 |
| Pascal p-value | 0.146 |
| DMG | 1 (# studies) |
| eGene | Cortex Myers' cis & trans |
| Support | CELL ADHESION AND TRANSSYNAPTIC SIGNALING |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.04359 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.04047 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg04416247 | 3 | 85213671 | CADM2 | 2.455E-4 | 0.511 | 0.037 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16894557 | chr6 | 28999825 | CADM2 | 253559 | 0.18 | trans |
Section II. Transcriptome annotation
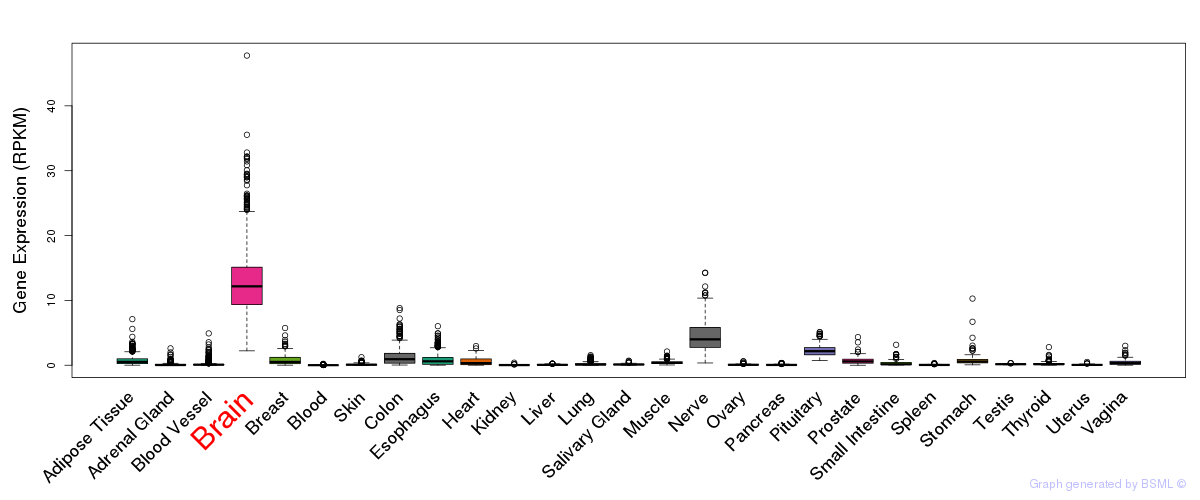
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME CELL CELL COMMUNICATION | 120 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME ADHERENS JUNCTIONS INTERACTIONS | 27 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CELL JUNCTION ORGANIZATION | 56 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL JUNCTION ORGANIZATION | 78 | 43 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS DN | 120 | 81 | All SZGR 2.0 genes in this pathway |
| ZHENG GLIOBLASTOMA PLASTICITY UP | 250 | 168 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE UP | 149 | 85 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE DN | 264 | 159 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-101 | 176 | 182 | 1A | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-124.1 | 261 | 267 | 1A | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-140 | 1180 | 1186 | 1A | hsa-miR-140brain | AGUGGUUUUACCCUAUGGUAG |
| miR-141/200a | 1974 | 1980 | 1A | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-143 | 277 | 283 | m8 | hsa-miR-143brain | UGAGAUGAAGCACUGUAGCUCA |
| miR-144 | 176 | 182 | 1A | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-153 | 1345 | 1351 | m8 | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-17-5p/20/93.mr/106/519.d | 1929 | 1935 | m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-182 | 1276 | 1283 | 1A,m8 | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-186 | 166 | 173 | 1A,m8 | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
| hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU | ||||
| miR-203.1 | 199 | 205 | m8 | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-21 | 1140 | 1146 | 1A | hsa-miR-21brain | UAGCUUAUCAGACUGAUGUUGA |
| hsa-miR-590 | GAGCUUAUUCAUAAAAGUGCAG | ||||
| miR-218 | 1111 | 1118 | 1A,m8 | hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU |
| miR-30-5p | 457 | 463 | 1A | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-338 | 39 | 46 | 1A,m8 | hsa-miR-338brain | UCCAGCAUCAGUGAUUUUGUUGA |
| miR-34b | 816 | 822 | 1A | hsa-miR-34b | UAGGCAGUGUCAUUAGCUGAUUG |
| miR-369-3p | 1006 | 1012 | 1A | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-374 | 1006 | 1013 | 1A,m8 | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-485-5p | 1159 | 1166 | 1A,m8 | hsa-miR-485-5p | AGAGGCUGGCCGUGAUGAAUUC |
| miR-495 | 1096 | 1102 | 1A | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| miR-543 | 565 | 571 | m8 | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
| hsa-miR-543 | AAACAUUCGCGGUGCACUUCU | ||||
| hsa-miR-543 | AAACAUUCGCGGUGCACUUCU | ||||
| miR-93.hd/291-3p/294/295/302/372/373/520 | 1928 | 1934 | m8 | hsa-miR-93brain | AAAGUGCUGUUCGUGCAGGUAG |
| hsa-miR-302a | UAAGUGCUUCCAUGUUUUGGUGA | ||||
| hsa-miR-302b | UAAGUGCUUCCAUGUUUUAGUAG | ||||
| hsa-miR-302c | UAAGUGCUUCCAUGUUUCAGUGG | ||||
| hsa-miR-302d | UAAGUGCUUCCAUGUUUGAGUGU | ||||
| hsa-miR-372 | AAAGUGCUGCGACAUUUGAGCGU | ||||
| hsa-miR-373 | GAAGUGCUUCGAUUUUGGGGUGU | ||||
| hsa-miR-520e | AAAGUGCUUCCUUUUUGAGGG | ||||
| hsa-miR-520a | AAAGUGCUUCCCUUUGGACUGU | ||||
| hsa-miR-520b | AAAGUGCUUCCUUUUAGAGGG | ||||
| hsa-miR-520c | AAAGUGCUUCCUUUUAGAGGGUU | ||||
| hsa-miR-520d | AAAGUGCUUCUCUUUGGUGGGUU | ||||
| miR-96 | 1277 | 1283 | 1A | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.