Gene Page: RAD54B
Summary ?
| GeneID | 25788 |
| Symbol | RAD54B |
| Synonyms | RDH54 |
| Description | RAD54 homolog B (S. cerevisiae) |
| Reference | MIM:604289|HGNC:HGNC:17228|Ensembl:ENSG00000197275|HPRD:05049|HPRD:09966|Vega:OTTHUMG00000133658 |
| Gene type | protein-coding |
| Map location | 8q22.1 |
| Pascal p-value | 0.816 |
| Fetal beta | 0.803 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg15995002 | 8 | 95556400 | RAD54B | -0.021 | 0.3 | DMG:Nishioka_2013 |
Section II. Transcriptome annotation
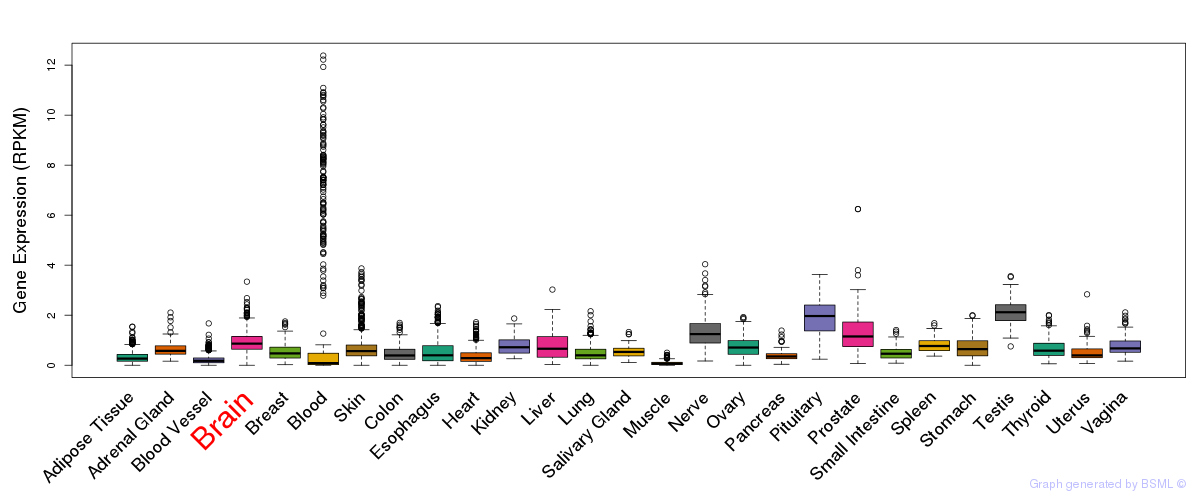
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KCTD7 | 0.96 | 0.95 |
| ZNF697 | 0.96 | 0.94 |
| SRC | 0.95 | 0.97 |
| LDOC1L | 0.95 | 0.97 |
| KDM4B | 0.95 | 0.96 |
| SUFU | 0.95 | 0.94 |
| XKR7 | 0.95 | 0.96 |
| SBK1 | 0.95 | 0.95 |
| RBM9 | 0.95 | 0.97 |
| DCAF7 | 0.95 | 0.95 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.70 | -0.80 |
| AF347015.31 | -0.68 | -0.92 |
| S100B | -0.68 | -0.87 |
| AF347015.27 | -0.68 | -0.90 |
| AIFM3 | -0.68 | -0.77 |
| HLA-F | -0.67 | -0.77 |
| HEPN1 | -0.67 | -0.77 |
| MT-CO2 | -0.67 | -0.92 |
| SERPINB6 | -0.67 | -0.75 |
| AF347015.33 | -0.66 | -0.89 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CCDC33 | FLJ23168 | FLJ32855 | MGC34145 | coiled-coil domain containing 33 | Two-hybrid | BioGRID | 16189514 |
| CD2BP2 | FWP010 | LIN1 | Snu40 | U5-52K | CD2 (cytoplasmic tail) binding protein 2 | Affinity Capture-MS | BioGRID | 17353931 |
| CDC23 | APC8 | cell division cycle 23 homolog (S. cerevisiae) | Two-hybrid | BioGRID | 16189514 |
| CSNK1E | HCKIE | MGC10398 | casein kinase 1, epsilon | Two-hybrid | BioGRID | 16189514 |
| DMC1 | DMC1H | HsLim15 | LIM15 | MGC150472 | MGC150473 | dJ199H16.1 | DMC1 dosage suppressor of mck1 homolog, meiosis-specific homologous recombination (yeast) | hDmc1 interacts with Rad54B. | BIND | 15164066 |
| DYRK2 | FLJ21217 | FLJ21365 | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 | Two-hybrid | BioGRID | 16189514 |
| GORASP2 | DKFZp434D156 | FLJ13139 | GOLPH6 | GRASP55 | GRS2 | p59 | golgi reassembly stacking protein 2, 55kDa | Two-hybrid | BioGRID | 16189514 |
| LNX1 | LNX | MPDZ | PDZRN2 | ligand of numb-protein X 1 | Two-hybrid | BioGRID | 16189514 |
| LOC170082 | MGC17403 | TFIIS central domain-containing protein 1 | Two-hybrid | BioGRID | 16189514 |
| MYO15B | FLJ21606 | FLJ22686 | FLJ40577 | FLJ44494 | MYO15BP | myosin XVB pseudogene | Two-hybrid | BioGRID | 16189514 |
| PSMA1 | HC2 | MGC14542 | MGC14575 | MGC14751 | MGC1667 | MGC21459 | MGC22853 | MGC23915 | NU | PROS30 | proteasome (prosome, macropain) subunit, alpha type, 1 | Two-hybrid | BioGRID | 16189514 |
| RAD51 | BRCC5 | HRAD51 | HsRad51 | HsT16930 | RAD51A | RECA | RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae) | - | HPRD,BioGRID | 10851248 |
| RAD54B | FSBP | RDH54 | RAD54 homolog B (S. cerevisiae) | Two-hybrid | BioGRID | 16189514 |
| RIBC2 | C22orf11 | RIB43A domain with coiled-coils 2 | Two-hybrid | BioGRID | 16189514 |
| TRAPPC6A | HSPC289 | MGC2650 | TRS33 | trafficking protein particle complex 6A | Two-hybrid | BioGRID | 16189514 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | Two-hybrid | BioGRID | 16189514 |
| ZNF580 | - | zinc finger protein 580 | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG HOMOLOGOUS RECOMBINATION | 28 | 17 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| SOTIRIOU BREAST CANCER GRADE 1 VS 3 UP | 151 | 84 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| DOANE RESPONSE TO ANDROGEN DN | 241 | 146 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY UP | 430 | 232 | All SZGR 2.0 genes in this pathway |
| ODONNELL TFRC TARGETS DN | 139 | 76 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR DN | 64 | 39 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR DN | 75 | 50 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER CLUSTER 2 | 33 | 17 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 DN | 156 | 106 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES 1 | 379 | 235 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH OCT4 TARGETS | 290 | 172 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH NOS TARGETS | 179 | 105 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 8Q12 Q22 AMPLICON | 132 | 82 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN DN | 271 | 175 | All SZGR 2.0 genes in this pathway |
| FUJII YBX1 TARGETS DN | 202 | 132 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE DN | 264 | 159 | All SZGR 2.0 genes in this pathway |
| RUIZ TNC TARGETS DN | 142 | 79 | All SZGR 2.0 genes in this pathway |
| KOBAYASHI EGFR SIGNALING 24HR DN | 251 | 151 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR UP | 324 | 193 | All SZGR 2.0 genes in this pathway |
| SMIRNOV RESPONSE TO IR 2HR DN | 55 | 35 | All SZGR 2.0 genes in this pathway |