Gene Page: LSM4
Summary ?
| GeneID | 25804 |
| Symbol | LSM4 |
| Synonyms | GRP|YER112W |
| Description | LSM4 homolog, U6 small nuclear RNA and mRNA degradation associated |
| Reference | MIM:607284|HGNC:HGNC:17259|Ensembl:ENSG00000130520|HPRD:06284|Vega:OTTHUMG00000183357 |
| Gene type | protein-coding |
| Map location | 19p13.11 |
| Pascal p-value | 0.248 |
| Sherlock p-value | 0.045 |
| Fetal beta | -0.302 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0833 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
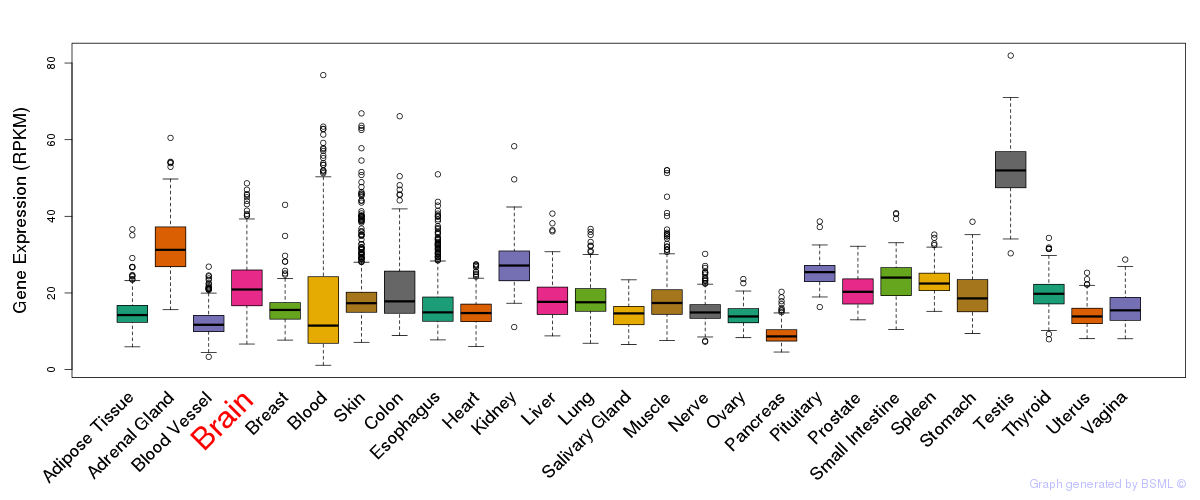
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003723 | RNA binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 15231747 |17353931 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006397 | mRNA processing | IEA | - | |
| GO:0008380 | RNA splicing | TAS | 10523320 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005688 | snRNP U6 | TAS | 10369684 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CALCOCO2 | MGC17318 | NDP52 | calcium binding and coiled-coil domain 2 | Two-hybrid | BioGRID | 16189514 |
| LSM1 | CASM | YJL124C | LSM1 homolog, U6 small nuclear RNA associated (S. cerevisiae) | - | HPRD,BioGRID | 11920408 |
| LSM2 | C6orf28 | G7b | YBL026W | snRNP | LSM2 homolog, U6 small nuclear RNA associated (S. cerevisiae) | - | HPRD,BioGRID | 11920408 |
| LSM3 | SMX4 | USS2 | YLR438C | LSM3 homolog, U6 small nuclear RNA associated (S. cerevisiae) | - | HPRD,BioGRID | 11920408 |
| LSM4 | YER112W | LSM4 homolog, U6 small nuclear RNA associated (S. cerevisiae) | Affinity Capture-Western | BioGRID | 11920408 |
| LSM5 | FLJ12710 | YER146W | LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) | - | HPRD | 11920408 |
| LSM6 | YDR378C | LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) | - | HPRD,BioGRID | 11920408 |
| LSM7 | YNL147W | LSM7 homolog, U6 small nuclear RNA associated (S. cerevisiae) | - | HPRD,BioGRID | 11920408 |
| LSM8 | YJR022W | LSM8 homolog, U6 small nuclear RNA associated (S. cerevisiae) | - | HPRD,BioGRID | 11920408 |
| NMI | - | N-myc (and STAT) interactor | - | HPRD,BioGRID | 15231747 |
| PLEKHA5 | FLJ10667 | FLJ31492 | KIAA1686 | PEPP2 | pleckstrin homology domain containing, family A member 5 | - | HPRD,BioGRID | 15231747 |
| SMN1 | BCD541 | SMA | SMA1 | SMA2 | SMA3 | SMA4 | SMA@ | SMN | SMNT | T-BCD541 | survival of motor neuron 1, telomeric | - | HPRD,BioGRID | 10851237 |
| SNRPE | B-raf | SME | small nuclear ribonucleoprotein polypeptide E | - | HPRD,BioGRID | 15231747 |
| XRN1 | DKFZp434P0721 | DKFZp686B22225 | DKFZp686F19113 | FLJ41903 | SEP1 | 5'-3' exoribonuclease 1 | Two-hybrid | BioGRID | 15231747 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG RNA DEGRADATION | 59 | 37 | All SZGR 2.0 genes in this pathway |
| KEGG SPLICEOSOME | 128 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | 15 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF MRNA | 284 | 128 | All SZGR 2.0 genes in this pathway |
| REACTOME DEADENYLATION DEPENDENT MRNA DECAY | 48 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF RNA | 330 | 155 | All SZGR 2.0 genes in this pathway |
| NAKAMURA CANCER MICROENVIRONMENT DN | 46 | 29 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING DN | 46 | 29 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 UP | 201 | 125 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS DN | 310 | 188 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN | 362 | 238 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR DN | 129 | 84 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR DN | 209 | 122 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF DN | 228 | 137 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| ROSTY CERVICAL CANCER PROLIFERATION CLUSTER | 140 | 73 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 120 MCF10A | 43 | 28 | All SZGR 2.0 genes in this pathway |
| MOREAUX B LYMPHOCYTE MATURATION BY TACI DN | 73 | 45 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI DN | 172 | 107 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| MODY HIPPOCAMPUS PRENATAL | 42 | 33 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 6HR UP | 71 | 48 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR UP | 294 | 199 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE UP | 212 | 128 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6 | 456 | 285 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| WONG EMBRYONIC STEM CELL CORE | 335 | 193 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 17 | 181 | 101 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR UP | 324 | 193 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |