Gene Page: GALC
Summary ?
| GeneID | 2581 |
| Symbol | GALC |
| Synonyms | - |
| Description | galactosylceramidase |
| Reference | MIM:606890|HGNC:HGNC:4115|Ensembl:ENSG00000054983|HPRD:06057|Vega:OTTHUMG00000028646 |
| Gene type | protein-coding |
| Map location | 14q31 |
| Pascal p-value | 0.578 |
| Sherlock p-value | 0.353 |
| Fetal beta | -0.532 |
| eGene | Cerebellar Hemisphere Cerebellum |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
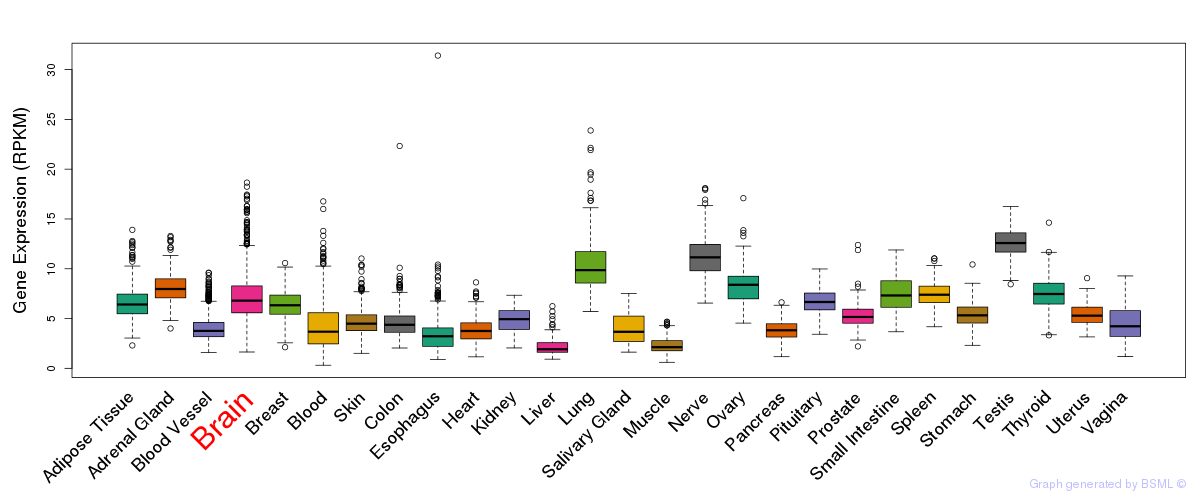
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PEBP1 | 0.83 | 0.82 |
| UBB | 0.81 | 0.84 |
| UQCRC1 | 0.81 | 0.81 |
| SNRPN | 0.80 | 0.80 |
| APEH | 0.79 | 0.75 |
| WDR45 | 0.78 | 0.76 |
| ATP6V0C | 0.78 | 0.79 |
| SLC25A11 | 0.77 | 0.80 |
| ABHD14A | 0.77 | 0.80 |
| RNF167 | 0.77 | 0.83 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| EIF5B | -0.51 | -0.61 |
| AC010300.1 | -0.48 | -0.56 |
| AC005921.3 | -0.47 | -0.62 |
| SFRS18 | -0.45 | -0.55 |
| NSBP1 | -0.43 | -0.42 |
| AF347015.18 | -0.41 | -0.25 |
| AF347015.21 | -0.40 | -0.23 |
| ANKRD36B | -0.39 | -0.40 |
| AP000769.2 | -0.39 | -0.40 |
| AC073534.1 | -0.39 | -0.53 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG SPHINGOLIPID METABOLISM | 40 | 26 | All SZGR 2.0 genes in this pathway |
| KEGG LYSOSOME | 121 | 83 | All SZGR 2.0 genes in this pathway |
| REACTOME GLYCOSPHINGOLIPID METABOLISM | 38 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME PHOSPHOLIPID METABOLISM | 198 | 112 | All SZGR 2.0 genes in this pathway |
| REACTOME SPHINGOLIPID METABOLISM | 69 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP | 457 | 269 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP | 633 | 376 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 3 4WK UP | 214 | 144 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL DN | 186 | 107 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL TGFB1 TARGETS UP | 169 | 127 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 2 | 127 | 92 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS UP | 292 | 168 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO DN | 200 | 112 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| ALCALAY AML BY NPM1 LOCALIZATION DN | 184 | 132 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED DN | 246 | 180 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| LIAN LIPA TARGETS 6M | 74 | 47 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS DN | 31 | 23 | All SZGR 2.0 genes in this pathway |
| MAHAJAN RESPONSE TO IL1A DN | 76 | 57 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| HOWLIN CITED1 TARGETS 1 DN | 37 | 22 | All SZGR 2.0 genes in this pathway |
| HOWLIN CITED1 TARGETS 2 DN | 17 | 11 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY UV | 62 | 43 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE UP | 226 | 164 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 DN | 315 | 201 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF DN | 235 | 144 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| VALK AML WITH CEBPA | 37 | 27 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING DN | 225 | 124 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS UP | 217 | 131 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS GROUP2 | 60 | 38 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 UP | 140 | 94 | All SZGR 2.0 genes in this pathway |
| KASLER HDAC7 TARGETS 1 UP | 194 | 133 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 7 | 403 | 240 | All SZGR 2.0 genes in this pathway |
| RATTENBACHER BOUND BY CELF1 | 467 | 251 | All SZGR 2.0 genes in this pathway |