Gene Page: PRKD2
Summary ?
| GeneID | 25865 |
| Symbol | PRKD2 |
| Synonyms | HSPC187|PKD2|nPKC-D2 |
| Description | protein kinase D2 |
| Reference | MIM:607074|HGNC:HGNC:17293|Ensembl:ENSG00000105287|HPRD:09523|Vega:OTTHUMG00000183431 |
| Gene type | protein-coding |
| Map location | 19q13.3 |
| Pascal p-value | 0.2 |
| Sherlock p-value | 0.494 |
| Fetal beta | 0.349 |
| DMG | 1 (# studies) |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cerebellum Cortex Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg27119358 | 19 | 47217696 | PRKD2 | 1.449E-4 | -0.498 | 0.031 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10282425 | chr7 | 129831082 | PRKD2 | 25865 | 0.08 | trans | ||
| rs12461848 | 19 | 47174893 | PRKD2 | ENSG00000105287.8 | 6.589E-10 | 0 | 45491 | gtex_brain_ba24 |
| rs874462 | 19 | 47176880 | PRKD2 | ENSG00000105287.8 | 6.516E-10 | 0 | 43504 | gtex_brain_ba24 |
| rs62134780 | 19 | 47188303 | PRKD2 | ENSG00000105287.8 | 6.532E-10 | 0 | 32081 | gtex_brain_ba24 |
| rs62134781 | 19 | 47188810 | PRKD2 | ENSG00000105287.8 | 6.516E-10 | 0 | 31574 | gtex_brain_ba24 |
| rs62134782 | 19 | 47205419 | PRKD2 | ENSG00000105287.8 | 6.321E-7 | 0 | 14965 | gtex_brain_ba24 |
| rs62134783 | 19 | 47206956 | PRKD2 | ENSG00000105287.8 | 3.814E-7 | 0 | 13428 | gtex_brain_ba24 |
| rs11083846 | 19 | 47207654 | PRKD2 | ENSG00000105287.8 | 2.431E-7 | 0 | 12730 | gtex_brain_ba24 |
| rs117579506 | 19 | 47209159 | PRKD2 | ENSG00000105287.8 | 3.88E-7 | 0 | 11225 | gtex_brain_ba24 |
| rs12461666 | 19 | 47217113 | PRKD2 | ENSG00000105287.8 | 4.144E-7 | 0 | 3271 | gtex_brain_ba24 |
| rs62134788 | 19 | 47217587 | PRKD2 | ENSG00000105287.8 | 4.167E-7 | 0 | 2797 | gtex_brain_ba24 |
| rs11668987 | 19 | 47227821 | PRKD2 | ENSG00000105287.8 | 7.579E-7 | 0 | -7437 | gtex_brain_ba24 |
| rs3215087 | 19 | 47230393 | PRKD2 | ENSG00000105287.8 | 4.57E-7 | 0 | -10009 | gtex_brain_ba24 |
| rs62134790 | 19 | 47237551 | PRKD2 | ENSG00000105287.8 | 4.071E-7 | 0 | -17167 | gtex_brain_ba24 |
| rs56374620 | 19 | 47241599 | PRKD2 | ENSG00000105287.8 | 3.514E-7 | 0 | -21215 | gtex_brain_ba24 |
| rs4802322 | 19 | 47242992 | PRKD2 | ENSG00000105287.8 | 1.608E-7 | 0 | -22608 | gtex_brain_ba24 |
| rs11668563 | 19 | 47243621 | PRKD2 | ENSG00000105287.8 | 2.611E-7 | 0 | -23237 | gtex_brain_ba24 |
| rs78887375 | 19 | 47247141 | PRKD2 | ENSG00000105287.8 | 1.823E-7 | 0 | -26757 | gtex_brain_ba24 |
| rs7258782 | 19 | 47247781 | PRKD2 | ENSG00000105287.8 | 1.669E-7 | 0 | -27397 | gtex_brain_ba24 |
| rs3786708 | 19 | 47248308 | PRKD2 | ENSG00000105287.8 | 2.023E-7 | 0 | -27924 | gtex_brain_ba24 |
| rs62134793 | 19 | 47248585 | PRKD2 | ENSG00000105287.8 | 1.663E-7 | 0 | -28201 | gtex_brain_ba24 |
| rs62134820 | 19 | 47256344 | PRKD2 | ENSG00000105287.8 | 3.586E-7 | 0 | -35960 | gtex_brain_ba24 |
| rs60104266 | 19 | 47257202 | PRKD2 | ENSG00000105287.8 | 9.667E-8 | 0 | -36818 | gtex_brain_ba24 |
| rs7249039 | 19 | 47266635 | PRKD2 | ENSG00000105287.8 | 8.294E-9 | 0 | -46251 | gtex_brain_ba24 |
| rs12459058 | 19 | 47274138 | PRKD2 | ENSG00000105287.8 | 3.608E-7 | 0 | -53754 | gtex_brain_ba24 |
| rs1644339 | 19 | 47281255 | PRKD2 | ENSG00000105287.8 | 8.244E-7 | 0 | -60871 | gtex_brain_ba24 |
Section II. Transcriptome annotation
General gene expression (GTEx)
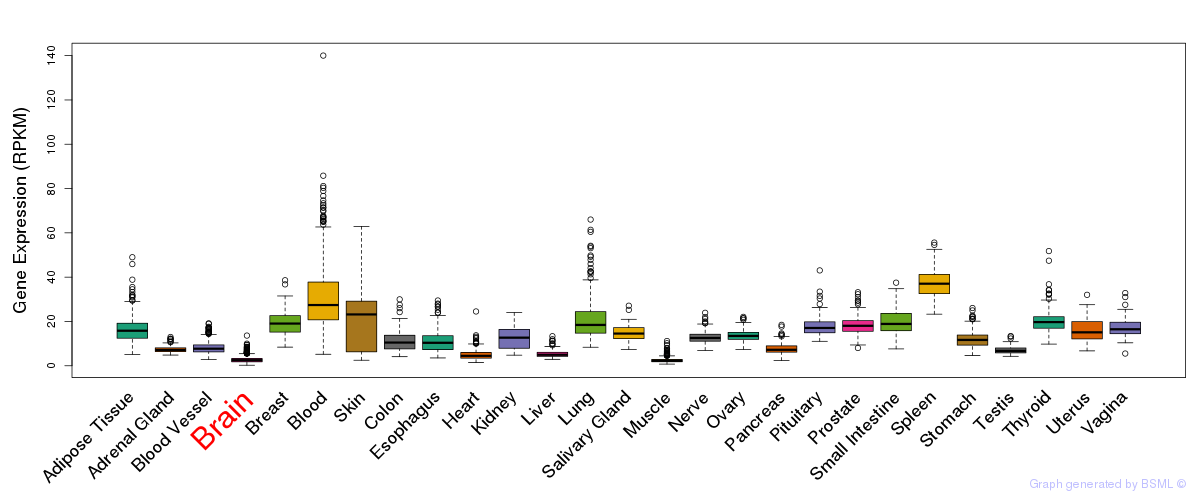
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL UP | 450 | 256 | All SZGR 2.0 genes in this pathway |
| ODONNELL TFRC TARGETS UP | 456 | 228 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| MAGRANGEAS MULTIPLE MYELOMA IGLL VS IGLK DN | 24 | 18 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS DN | 105 | 63 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY GAMMA AND UV RADIATION | 88 | 65 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE DN | 195 | 135 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| FIRESTEIN PROLIFERATION | 175 | 125 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER UP | 298 | 174 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 10 | 33 | 22 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S1 | 237 | 159 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA CLASSICAL | 162 | 122 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS DN | 213 | 127 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |