Gene Page: GAMT
Summary ?
| GeneID | 2593 |
| Symbol | GAMT |
| Synonyms | CCDS2|HEL-S-20|PIG2|TP53I2 |
| Description | guanidinoacetate N-methyltransferase |
| Reference | MIM:601240|HGNC:HGNC:4136|Ensembl:ENSG00000130005|HPRD:03142|Vega:OTTHUMG00000180095 |
| Gene type | protein-coding |
| Map location | 19p13.3 |
| Pascal p-value | 0.014 |
| Sherlock p-value | 0.748 |
| DEG p-value | DEG:Sanders_2014:DS1_p=0.149:DS1_beta=0.039100:DS2_p=1.64e-07:DS2_beta=0.265:DS2_FDR=3.06e-05 |
| Fetal beta | -0.647 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DEG:Sanders_2013 | Microarray | Whole-genome gene expression profiles using microarrays on lymphoblastoid cell lines (LCLs) from 413 cases and 446 controls. |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7830259 | chr8 | 14087643 | GAMT | 2593 | 0.01 | trans | ||
| rs7997312 | chr13 | 84518989 | GAMT | 2593 | 0.07 | trans |
Section II. Transcriptome annotation
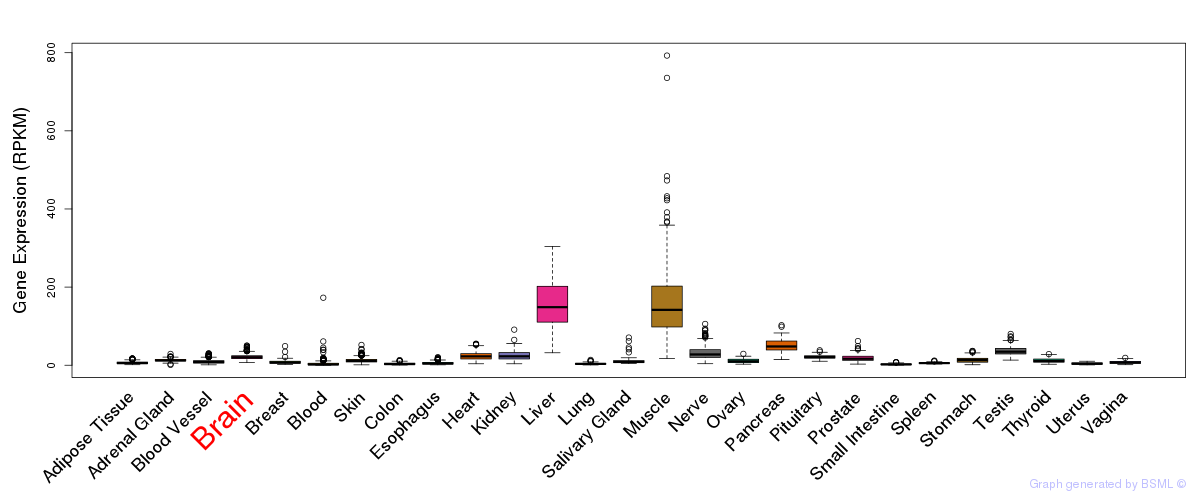
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG GLYCINE SERINE AND THREONINE METABOLISM | 31 | 26 | All SZGR 2.0 genes in this pathway |
| KEGG ARGININE AND PROLINE METABOLISM | 54 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | 200 | 136 | All SZGR 2.0 genes in this pathway |
| PUIFFE INVASION INHIBITED BY ASCITES DN | 145 | 91 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL UP | 380 | 215 | All SZGR 2.0 genes in this pathway |
| DOANE BREAST CANCER ESR1 UP | 112 | 72 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION CDC25 DN | 153 | 100 | All SZGR 2.0 genes in this pathway |
| OUELLET OVARIAN CANCER INVASIVE VS LMP DN | 11 | 8 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER DN | 203 | 134 | All SZGR 2.0 genes in this pathway |
| SMITH TERT TARGETS DN | 87 | 69 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| ALCALAY AML BY NPM1 LOCALIZATION DN | 184 | 132 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN | 287 | 208 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 48HR DN | 161 | 105 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| MATZUK SPERMATOZOA | 114 | 77 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G3 DN | 51 | 32 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS PROLIFERATION DN | 179 | 97 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT DN | 222 | 141 | All SZGR 2.0 genes in this pathway |
| KASLER HDAC7 TARGETS 2 DN | 32 | 23 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR DN | 244 | 157 | All SZGR 2.0 genes in this pathway |
| SMIRNOV RESPONSE TO IR 6HR UP | 166 | 97 | All SZGR 2.0 genes in this pathway |