Gene Page: GAPDH
Summary ?
| GeneID | 2597 |
| Symbol | GAPDH |
| Synonyms | G3PD|GAPD|HEL-S-162eP |
| Description | glyceraldehyde-3-phosphate dehydrogenase |
| Reference | MIM:138400|HGNC:HGNC:4141|Ensembl:ENSG00000111640|HPRD:00713|Vega:OTTHUMG00000137379 |
| Gene type | protein-coding |
| Map location | 12p13 |
| Pascal p-value | 0.733 |
| Sherlock p-value | 0.995 |
| Fetal beta | -0.26 |
| DMG | 1 (# studies) |
| eGene | Frontal Cortex BA9 |
| Support | CELL METABOLISM G2Cdb.human_mitochondria G2Cdb.humanNRC G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.6677 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg20917484 | 12 | 6644066 | GAPDH | 4.748E-4 | -0.237 | 0.046 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
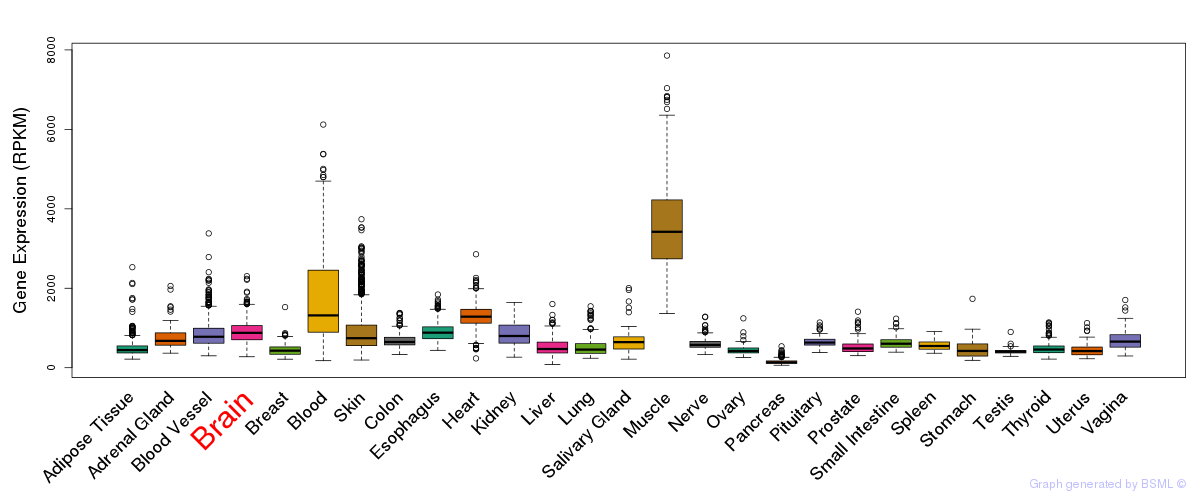
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003824 | catalytic activity | IEA | - | |
| GO:0005488 | binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 17540579 | |
| GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (phosphorylating) activity | IEA | - | |
| GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (phosphorylating) activity | NAS | 3170585 |7030790 | |
| GO:0016491 | oxidoreductase activity | IEA | - | |
| GO:0051287 | NAD binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006096 | glycolysis | NAS | - | |
| GO:0006006 | glucose metabolic process | IEA | - | |
| GO:0008152 | metabolic process | IEA | - | |
| GO:0055114 | oxidation reduction | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | NAS | - | |
| GO:0016020 | membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| APP | AAA | ABETA | ABPP | AD1 | APPI | CTFgamma | CVAP | PN2 | amyloid beta (A4) precursor protein | - | HPRD | 8473906 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | - | HPRD,BioGRID | 8872471 |
| ARL15 | ARFRP2 | FLJ20051 | ADP-ribosylation factor-like 15 | Two-hybrid | BioGRID | 16169070 |
| ATXN1 | ATX1 | D6S504E | SCA1 | ataxin 1 | - | HPRD,BioGRID | 8872471 |
| BPGM | - | 2,3-bisphosphoglycerate mutase | - | HPRD | 9308888 |
| BTBD2 | - | BTB (POZ) domain containing 2 | Two-hybrid | BioGRID | 16169070 |
| DYNLL1 | DLC1 | DLC8 | DNCL1 | DNCLC1 | LC8 | LC8a | MGC126137 | MGC126138 | PIN | hdlc1 | dynein, light chain, LC8-type 1 | - | HPRD | 14760703 |
| GAPDH | G3PD | GAPD | MGC88685 | glyceraldehyde-3-phosphate dehydrogenase | - | HPRD | 10617673 |
| GRM1 | GPRC1A | GRM1A | MGLUR1 | MGLUR1A | mGlu1 | glutamate receptor, metabotropic 1 | - | HPRD | 9886087 |
| HBXIP | MGC71071 | XIP | hepatitis B virus x interacting protein | Two-hybrid | BioGRID | 16169070 |
| HTT | HD | IT15 | huntingtin | - | HPRD,BioGRID | 8612237 |
| LAMA4 | DKFZp686D23145 | LAMA3 | LAMA4*-1 | laminin, alpha 4 | Two-hybrid | BioGRID | 16169070 |
| NPAT | E14 | nuclear protein, ataxia-telangiectasia locus | Affinity Capture-Western | BioGRID | 12887926 |
| PAFAH1B3 | - | platelet-activating factor acetylhydrolase, isoform Ib, gamma subunit 29kDa | Two-hybrid | BioGRID | 16169070 |
| PCDHA4 | CNR1 | CNRN1 | CRNR1 | MGC138307 | MGC142169 | PCDH-ALPHA4 | protocadherin alpha 4 | Two-hybrid | BioGRID | 16169070 |
| PDIA2 | PDA2 | PDI | PDIP | PDIR | protein disulfide isomerase family A, member 2 | - | HPRD,BioGRID | 11488911 |
| PGK1 | MGC117307 | MGC142128 | MGC8947 | MIG10 | PGKA | phosphoglycerate kinase 1 | - | HPRD,BioGRID | 8075252 |
| PLD2 | - | phospholipase D2 | - | HPRD,BioGRID | 12753082 |
| POU2F1 | OCT1 | OTF1 | POU class 2 homeobox 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 12887926 |
| POU2F2 | OCT2 | OTF2 | Oct-2 | POU class 2 homeobox 2 | Reconstituted Complex | BioGRID | 12887926 |
| PRDX1 | MSP23 | NKEFA | PAG | PAGA | PAGB | PRX1 | PRXI | TDPX2 | peroxiredoxin 1 | Two-hybrid | BioGRID | 16169070 |
| PRKCI | DXS1179E | MGC26534 | PKCI | nPKC-iota | protein kinase C, iota | - | HPRD,BioGRID | 11724794 |
| PRKG1 | CGKI | DKFZp686K042 | FLJ36117 | MGC71944 | PGK | PKG | PRKG1B | PRKGR1B | cGKI-BETA | cGKI-alpha | protein kinase, cGMP-dependent, type I | Affinity Capture-Western | BioGRID | 9308888 |
| PSMD11 | MGC3844 | Rpn6 | S9 | p44.5 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 | Two-hybrid | BioGRID | 16169070 |
| QRICH2 | DKFZp434P0316 | glutamine rich 2 | Affinity Capture-MS | BioGRID | 17353931 |
| RPA2 | REPA2 | RPA32 | replication protein A2, 32kDa | Two-hybrid | BioGRID | 16169070 |
| S100A6 | 2A9 | 5B10 | CABP | CACY | PRA | S100 calcium binding protein A6 | - | HPRD | 1999197 |
| SERPINB9 | CAP-3 | CAP3 | PI9 | serpin peptidase inhibitor, clade B (ovalbumin), member 9 | Two-hybrid | BioGRID | 16169070 |
| SLC2A1 | DYT17 | DYT18 | GLUT | GLUT1 | MGC141895 | MGC141896 | PED | solute carrier family 2 (facilitated glucose transporter), member 1 | - | HPRD,BioGRID | 2394733 |
| STIP1 | HOP | IEF-SSP-3521 | P60 | STI1 | STI1L | stress-induced-phosphoprotein 1 | Affinity Capture-Western | BioGRID | 12887926 |
| SYNCRIP | GRY-RBP | HNRPQ1 | NSAP1 | RP1-3J17.2 | dJ3J17.2 | hnRNP-Q | pp68 | synaptotagmin binding, cytoplasmic RNA interacting protein | NSAP-1 interacts with GAPDH. | BIND | 15479637 |
| TK1 | TK2 | thymidine kinase 1, soluble | Two-hybrid | BioGRID | 16169070 |
| TXN | DKFZp686B1993 | MGC61975 | TRX | TRX1 | thioredoxin | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG GLYCOLYSIS GLUCONEOGENESIS | 62 | 41 | All SZGR 2.0 genes in this pathway |
| KEGG ALZHEIMERS DISEASE | 169 | 110 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MTA3 PATHWAY | 19 | 10 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GLYCOLYSIS PATHWAY | 10 | 9 | All SZGR 2.0 genes in this pathway |
| WNT SIGNALING | 89 | 71 | All SZGR 2.0 genes in this pathway |
| PID MYC ACTIV PATHWAY | 79 | 62 | All SZGR 2.0 genes in this pathway |
| REACTOME GLYCOLYSIS | 29 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUCONEOGENESIS | 34 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF CARBOHYDRATES | 247 | 154 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUCOSE METABOLISM | 69 | 42 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA UP | 92 | 57 | All SZGR 2.0 genes in this pathway |
| KORKOLA EMBRYONAL CARCINOMA UP | 41 | 27 | All SZGR 2.0 genes in this pathway |
| KORKOLA YOLK SAC TUMOR UP | 20 | 14 | All SZGR 2.0 genes in this pathway |
| KORKOLA CHORIOCARCINOMA UP | 6 | 5 | All SZGR 2.0 genes in this pathway |
| KORKOLA SEMINOMA UP | 44 | 27 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA DN | 116 | 79 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN | 362 | 238 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 2HR UP | 27 | 19 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 6HR UP | 55 | 34 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS CDC25 UP | 58 | 39 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| DACOSTA LOW DOSE UV RESPONSE VIA ERCC3 XPCS DN | 9 | 6 | All SZGR 2.0 genes in this pathway |
| DIRMEIER LMP1 RESPONSE LATE DN | 32 | 22 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| ALCALA APOPTOSIS | 88 | 60 | All SZGR 2.0 genes in this pathway |
| GROSS HIF1A TARGETS DN | 25 | 15 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA HIF1A DN | 110 | 78 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| MORI PRE BI LYMPHOCYTE UP | 80 | 54 | All SZGR 2.0 genes in this pathway |
| MORI SMALL PRE BII LYMPHOCYTE DN | 76 | 52 | All SZGR 2.0 genes in this pathway |
| SHIPP DLBCL CURED VS FATAL DN | 45 | 30 | All SZGR 2.0 genes in this pathway |
| GNATENKO PLATELET SIGNATURE | 48 | 28 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK UP | 87 | 58 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT REJECTED VS OK DN | 51 | 35 | All SZGR 2.0 genes in this pathway |
| GALINDO IMMUNE RESPONSE TO ENTEROTOXIN | 85 | 67 | All SZGR 2.0 genes in this pathway |
| FERRANDO T ALL WITH MLL ENL FUSION DN | 87 | 57 | All SZGR 2.0 genes in this pathway |
| MUNSHI MULTIPLE MYELOMA UP | 81 | 52 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION | 405 | 264 | All SZGR 2.0 genes in this pathway |
| TRAYNOR RETT SYNDROM UP | 45 | 33 | All SZGR 2.0 genes in this pathway |
| BRACHAT RESPONSE TO METHOTREXATE DN | 27 | 19 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| HARRIS HYPOXIA | 81 | 64 | All SZGR 2.0 genes in this pathway |
| NATSUME RESPONSE TO INTERFERON BETA DN | 52 | 33 | All SZGR 2.0 genes in this pathway |
| MACLACHLAN BRCA1 TARGETS UP | 21 | 16 | All SZGR 2.0 genes in this pathway |
| BRACHAT RESPONSE TO CAMPTOTHECIN DN | 46 | 31 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 6HR UP | 71 | 48 | All SZGR 2.0 genes in this pathway |
| SEMENZA HIF1 TARGETS | 36 | 32 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND ENDOTHELIUM | 66 | 47 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| DAIRKEE CANCER PRONE RESPONSE BPA E2 | 118 | 65 | All SZGR 2.0 genes in this pathway |
| HONMA DOCETAXEL RESISTANCE | 34 | 23 | All SZGR 2.0 genes in this pathway |
| WHITEFORD PEDIATRIC CANCER MARKERS | 116 | 63 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR UP | 294 | 199 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA METAGENE | 242 | 168 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC | 420 | 269 | All SZGR 2.0 genes in this pathway |
| GOLDRATH ANTIGEN RESPONSE | 346 | 192 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 DN | 240 | 153 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6 | 456 | 285 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| NGO MALIGNANT GLIOMA 1P LOH | 17 | 13 | All SZGR 2.0 genes in this pathway |
| DANG MYC TARGETS UP | 143 | 100 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS UP | 221 | 135 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 14 | 143 | 86 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 16 | 79 | 47 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| GREGORY SYNTHETIC LETHAL WITH IMATINIB | 145 | 83 | All SZGR 2.0 genes in this pathway |
| WANG ADIPOGENIC GENES REPRESSED BY SIRT1 | 28 | 21 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP D | 280 | 158 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| PECE MAMMARY STEM CELL UP | 146 | 75 | All SZGR 2.0 genes in this pathway |
| HOLLEMAN PREDNISOLONE RESISTANCE B ALL DN | 12 | 8 | All SZGR 2.0 genes in this pathway |