Gene Page: IBTK
Summary ?
| GeneID | 25998 |
| Symbol | IBTK |
| Synonyms | BTBD26|BTKI |
| Description | inhibitor of Bruton tyrosine kinase |
| Reference | MIM:606457|HGNC:HGNC:17853|Ensembl:ENSG00000005700|HPRD:12105|Vega:OTTHUMG00000015102 |
| Gene type | protein-coding |
| Map location | 6q14.1 |
| Pascal p-value | 0.107 |
| DMG | 1 (# studies) |
| eGene | Frontal Cortex BA9 Putamen basal ganglia |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg07596845 | 6 | 82957841 | IBTK | 5.55E-10 | -0.014 | 8.9E-7 | DMG:Jaffe_2016 |
| cg10797726 | 6 | 82957093 | IBTK | 1.94E-8 | -0.009 | 6.81E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs477698 | 6 | 82975248 | IBTK | ENSG00000005700.10 | 3.52627E-6 | 0.05 | -17777 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
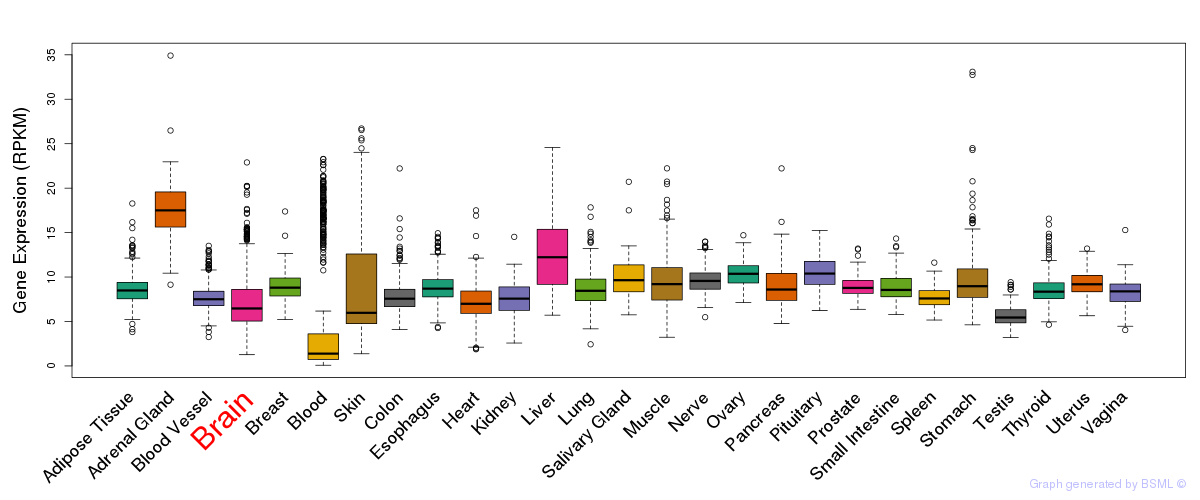
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ARFGEF2 | 0.87 | 0.88 |
| KIAA0513 | 0.87 | 0.86 |
| MFSD6 | 0.86 | 0.87 |
| AP006216.1 | 0.86 | 0.89 |
| STX1B | 0.86 | 0.83 |
| SCN8A | 0.85 | 0.87 |
| AAK1 | 0.85 | 0.83 |
| ATP2B3 | 0.85 | 0.84 |
| MBNL1 | 0.85 | 0.81 |
| TTLL7 | 0.85 | 0.86 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DBI | -0.51 | -0.59 |
| C1orf61 | -0.51 | -0.59 |
| RPL35 | -0.50 | -0.57 |
| RHOC | -0.49 | -0.55 |
| GNG11 | -0.48 | -0.53 |
| AC006276.2 | -0.47 | -0.50 |
| RPS7 | -0.47 | -0.45 |
| BCL7C | -0.46 | -0.51 |
| MYL12A | -0.46 | -0.53 |
| BTBD17 | -0.46 | -0.56 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 11577348 |17353931 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0016020 | membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID BCR 5PATHWAY | 65 | 50 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA PROGENITOR DN | 66 | 42 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS TOP50 UP | 38 | 27 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS DN | 384 | 230 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| ZHENG FOXP3 TARGETS IN THYMUS UP | 196 | 137 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| HOFFMANN IMMATURE TO MATURE B LYMPHOCYTE UP | 43 | 34 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS UP | 221 | 135 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| CHANGOLKAR H2AFY TARGETS UP | 48 | 28 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL DN | 428 | 246 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-25/32/92/363/367 | 1157 | 1164 | 1A,m8 | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.