Gene Page: ERAL1
Summary ?
| GeneID | 26284 |
| Symbol | ERAL1 |
| Synonyms | CEGA|ERA|ERA-W|ERAL1A|H-ERA|HERA-A|HERA-B |
| Description | Era-like 12S mitochondrial rRNA chaperone 1 |
| Reference | MIM:607435|HGNC:HGNC:3424|Ensembl:ENSG00000132591|HPRD:08466|Vega:OTTHUMG00000132675 |
| Gene type | protein-coding |
| Map location | 17q11.2 |
| Pascal p-value | 0.246 |
| Sherlock p-value | 0.363 |
| Fetal beta | 0.355 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg07776409 | 17 | 27182127 | ERAL1 | 5.12E-9 | -0.013 | 2.9E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2070265 | chr17 | 27075422 | ERAL1 | 26284 | 0.08 | cis | ||
| rs4794836 | chr17 | 27082709 | ERAL1 | 26284 | 0.05 | cis | ||
| rs4795457 | chr17 | 27087928 | ERAL1 | 26284 | 0.05 | cis | ||
| rs7215310 | chr17 | 27088625 | ERAL1 | 26284 | 0.11 | cis | ||
| rs11658884 | chr17 | 27132741 | ERAL1 | 26284 | 0.04 | cis | ||
| rs4795465 | chr17 | 27151497 | ERAL1 | 26284 | 0.13 | cis | ||
| rs2242345 | chr17 | 27185826 | ERAL1 | 26284 | 0.07 | cis | ||
| rs16965255 | chr17 | 28182193 | ERAL1 | 26284 | 0.2 | cis | ||
| rs17762315 | chr5 | 76807576 | ERAL1 | 26284 | 0.02 | trans |
Section II. Transcriptome annotation
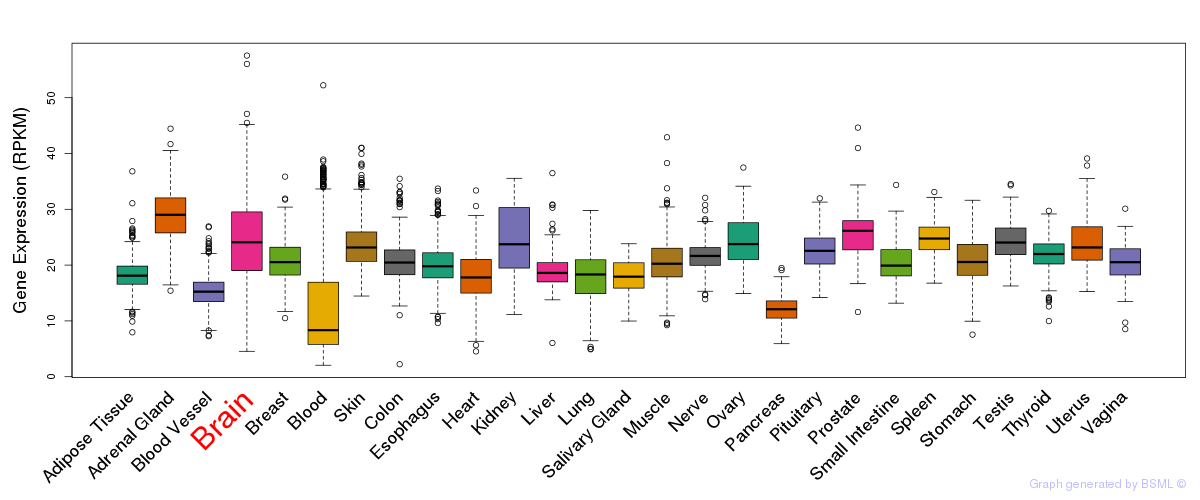
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| LAIHO COLORECTAL CANCER SERRATED DN | 86 | 47 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION DN | 234 | 147 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK DN | 196 | 131 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 5 6WK UP | 116 | 68 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 17Q11 Q21 AMPLICON | 133 | 78 | All SZGR 2.0 genes in this pathway |
| PENG GLUCOSE DEPRIVATION DN | 169 | 112 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC DN | 228 | 146 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| OUYANG PROSTATE CANCER PROGRESSION DN | 21 | 16 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |