Gene Page: GCK
Summary ?
| GeneID | 2645 |
| Symbol | GCK |
| Synonyms | FGQTL3|GK|GLK|HHF3|HK4|HKIV|HXKP|LGLK|MODY2 |
| Description | glucokinase |
| Reference | MIM:138079|HGNC:HGNC:4195|Ensembl:ENSG00000106633|HPRD:00680|Vega:OTTHUMG00000022903 |
| Gene type | protein-coding |
| Map location | 7p15.3-p15.1 |
| Pascal p-value | 0.016 |
| Fetal beta | -0.146 |
| DMG | 1 (# studies) |
| eGene | Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg11194698 | 7 | 44184840 | GCK | 5.323E-4 | 0.439 | 0.048 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
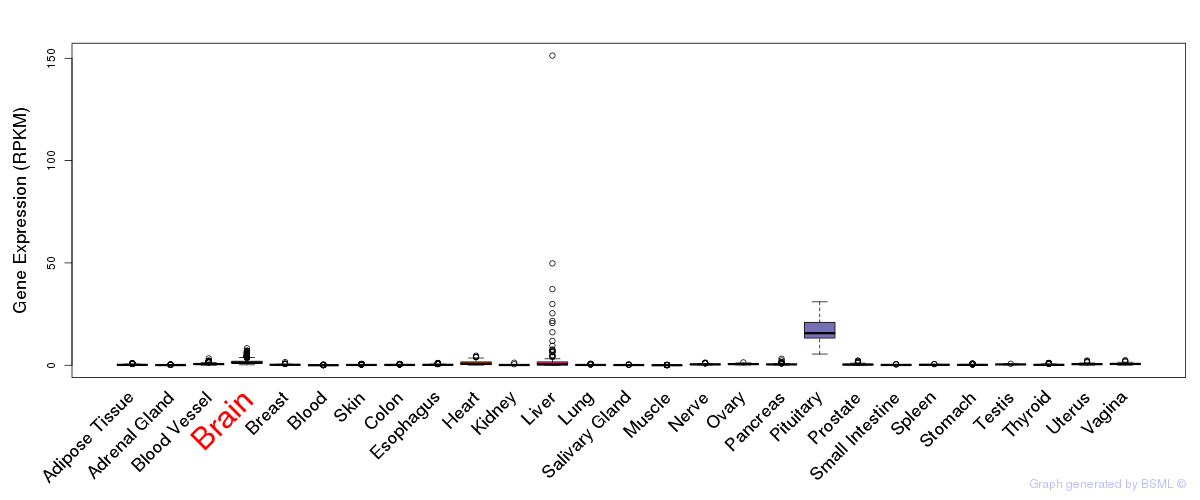
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ZNF135 | 0.82 | 0.75 |
| LMBR1L | 0.81 | 0.75 |
| ANKRD10 | 0.81 | 0.77 |
| CUL7 | 0.80 | 0.73 |
| PHKA2 | 0.80 | 0.75 |
| ZNF509 | 0.80 | 0.75 |
| ZNF140 | 0.80 | 0.77 |
| DFFB | 0.80 | 0.76 |
| ATRIP | 0.80 | 0.75 |
| ANKZF1 | 0.80 | 0.73 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.27 | -0.58 | -0.68 |
| FBXO2 | -0.57 | -0.61 |
| AIFM3 | -0.57 | -0.61 |
| MT-CO2 | -0.57 | -0.67 |
| HLA-F | -0.56 | -0.59 |
| MUSTN1 | -0.56 | -0.65 |
| TINAGL1 | -0.56 | -0.61 |
| AF347015.31 | -0.56 | -0.66 |
| AF347015.8 | -0.56 | -0.69 |
| ABCG2 | -0.56 | -0.58 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG GLYCOLYSIS GLUCONEOGENESIS | 62 | 41 | All SZGR 2.0 genes in this pathway |
| KEGG GALACTOSE METABOLISM | 26 | 22 | All SZGR 2.0 genes in this pathway |
| KEGG STARCH AND SUCROSE METABOLISM | 52 | 33 | All SZGR 2.0 genes in this pathway |
| KEGG AMINO SUGAR AND NUCLEOTIDE SUGAR METABOLISM | 44 | 30 | All SZGR 2.0 genes in this pathway |
| KEGG INSULIN SIGNALING PATHWAY | 137 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG TYPE II DIABETES MELLITUS | 47 | 41 | All SZGR 2.0 genes in this pathway |
| KEGG MATURITY ONSET DIABETES OF THE YOUNG | 25 | 19 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CHREBP2 PATHWAY | 42 | 35 | All SZGR 2.0 genes in this pathway |
| ST JNK MAPK PATHWAY | 40 | 30 | All SZGR 2.0 genes in this pathway |
| PID HNF3B PATHWAY | 45 | 37 | All SZGR 2.0 genes in this pathway |
| PID HIF1 TFPATHWAY | 66 | 52 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF BETA CELL DEVELOPMENT | 30 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | 20 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | 413 | 270 | All SZGR 2.0 genes in this pathway |
| REACTOME SLC MEDIATED TRANSMEMBRANE TRANSPORT | 241 | 157 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUCOSE TRANSPORT | 38 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF CARBOHYDRATES | 247 | 154 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | 27 | 19 | All SZGR 2.0 genes in this pathway |
| GALLUZZI PERMEABILIZE MITOCHONDRIA | 43 | 31 | All SZGR 2.0 genes in this pathway |
| LEE AGING CEREBELLUM UP | 84 | 58 | All SZGR 2.0 genes in this pathway |
| WENG POR TARGETS GLOBAL UP | 20 | 14 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS POLYSOMY7 UP | 79 | 50 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS LCP WITH H3K4ME3 | 174 | 100 | All SZGR 2.0 genes in this pathway |
| DUAN PRDM5 TARGETS | 79 | 52 | All SZGR 2.0 genes in this pathway |
| GUO TARGETS OF IRS1 AND IRS2 | 98 | 67 | All SZGR 2.0 genes in this pathway |