Gene Page: PABPC1
Summary ?
| GeneID | 26986 |
| Symbol | PABPC1 |
| Synonyms | PAB1|PABP|PABP1|PABPC2|PABPL1 |
| Description | poly(A) binding protein, cytoplasmic 1 |
| Reference | MIM:604679|HGNC:HGNC:8554|Ensembl:ENSG00000070756|HPRD:05247|Vega:OTTHUMG00000164779 |
| Gene type | protein-coding |
| Map location | 8q22.2-q23 |
| Pascal p-value | 0.626 |
| Sherlock p-value | 0.535 |
| Fetal beta | 0.849 |
| DMG | 1 (# studies) |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg05950485 | 8 | 101733214 | PABPC1 | 8.95E-9 | -0.007 | 4.09E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
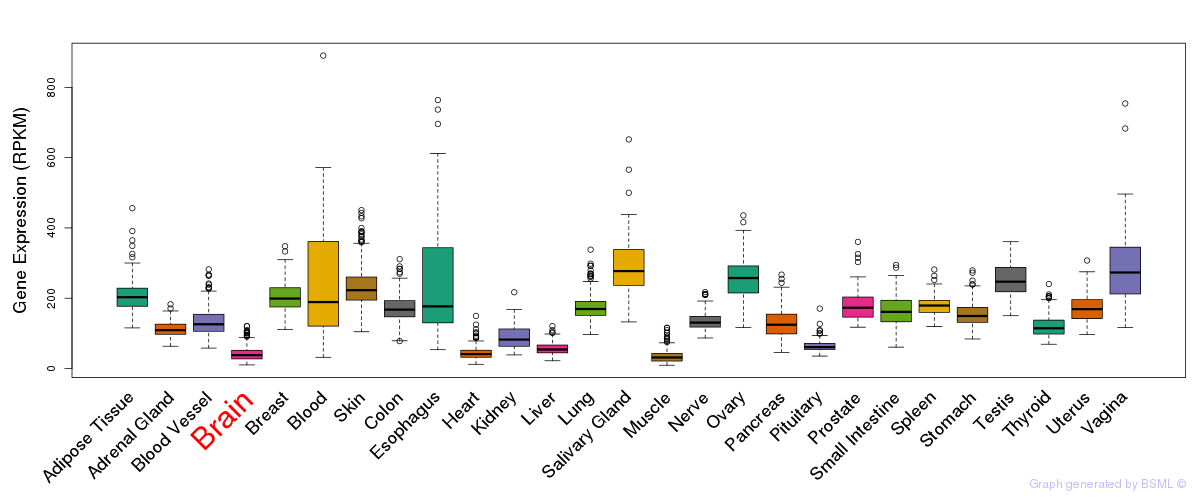
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| THADA | 0.93 | 0.93 |
| INTS7 | 0.93 | 0.93 |
| MCPH1 | 0.92 | 0.92 |
| DHX15 | 0.92 | 0.93 |
| RBBP4 | 0.92 | 0.91 |
| ZNF143 | 0.91 | 0.89 |
| XRN2 | 0.91 | 0.92 |
| KLHL7 | 0.91 | 0.92 |
| RBMX | 0.91 | 0.90 |
| TEX10 | 0.91 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.68 | -0.86 |
| AF347015.27 | -0.68 | -0.83 |
| AF347015.33 | -0.67 | -0.85 |
| FXYD1 | -0.67 | -0.85 |
| AF347015.31 | -0.67 | -0.83 |
| HLA-F | -0.67 | -0.76 |
| C5orf53 | -0.66 | -0.72 |
| AF347015.8 | -0.66 | -0.85 |
| IFI27 | -0.65 | -0.83 |
| MT-CYB | -0.65 | -0.82 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ANAPC5 | APC5 | anaphase promoting complex subunit 5 | - | HPRD,BioGRID | 15082755 |
| BCAR3 | KIAA0554 | NSP2 | SH2D3B | breast cancer anti-estrogen resistance 3 | Affinity Capture-MS | BioGRID | 17353931 |
| CNOT7 | CAF1 | hCAF-1 | CCR4-NOT transcription complex, subunit 7 | Affinity Capture-Western Reconstituted Complex | BioGRID | 18056425 |
| DYNLL1 | DLC1 | DLC8 | DNCL1 | DNCLC1 | LC8 | LC8a | MGC126137 | MGC126138 | PIN | hdlc1 | dynein, light chain, LC8-type 1 | Affinity Capture-MS | BioGRID | 17353931 |
| EIF4A1 | DDX2A | EIF-4A | EIF4A | eukaryotic translation initiation factor 4A, isoform 1 | Affinity Capture-Western | BioGRID | 9548260 |
| EIF4A2 | BM-010 | DDX2B | EIF4A | EIF4F | eukaryotic translation initiation factor 4A, isoform 2 | Affinity Capture-MS | BioGRID | 17353931 |
| EIF4B | EIF-4B | PRO1843 | eukaryotic translation initiation factor 4B | - | HPRD,BioGRID | 11274152 |
| EIF4G1 | DKFZp686A1451 | EIF4F | EIF4G | p220 | eukaryotic translation initiation factor 4 gamma, 1 | - | HPRD,BioGRID | 9857202 |
| ETF1 | D5S1995 | ERF | ERF1 | MGC111066 | RF1 | SUP45L1 | TB3-1 | eukaryotic translation termination factor 1 | Affinity Capture-Western | BioGRID | 18056425 |
| GNB2L1 | Gnb2-rs1 | H12.3 | HLC-7 | PIG21 | RACK1 | guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 | - | HPRD,BioGRID | 12388589 |
| GSPT1 | 551G9.2 | ETF3A | FLJ38048 | FLJ39067 | GST1 | eRF3a | G1 to S phase transition 1 | - | HPRD,BioGRID | 10358005 |
| GSPT2 | FLJ10441 | GST2 | eRF3b | G1 to S phase transition 2 | - | HPRD,BioGRID | 10358005 |
| IKBKE | IKK-i | IKKE | IKKI | KIAA0151 | MGC125294 | MGC125295 | MGC125297 | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon | Affinity Capture-MS | BioGRID | 17353931 |
| PAIP1 | MGC12360 | poly(A) binding protein interacting protein 1 | - | HPRD,BioGRID | 9548260 |11997512 |
| PAIP2 | MGC72018 | PAIP2A | poly(A) binding protein interacting protein 2 | - | HPRD,BioGRID | 11438674 |
| PAN2 | FLJ39360 | KIAA0710 | USP52 | hPAN2 | PAN2 polyA specific ribonuclease subunit homolog (S. cerevisiae) | - | HPRD,BioGRID | 14583602 |
| PAN3 | - | PAN3 polyA specific ribonuclease subunit homolog (S. cerevisiae) | Affinity Capture-Western Reconstituted Complex | BioGRID | 18056425 |
| PINX1 | FLJ20565 | LPTL | LPTS | MGC8850 | PIN2-interacting protein 1 | Affinity Capture-MS | BioGRID | 17353931 |
| PUF60 | FIR | FLJ31379 | RoBPI | SIAHBP1 | poly-U binding splicing factor 60KDa | Affinity Capture-MS | BioGRID | 17353931 |
| PXN | FLJ16691 | paxillin | - | HPRD,BioGRID | 11704675 |
| SFRS12 | DKFZp564B176 | MGC133045 | SRrp508 | SRrp86 | splicing factor, arginine/serine-rich 12 | - | HPRD | 14559993 |
| TOB1 | APRO6 | MGC104792 | MGC34446 | PIG49 | TOB | TROB | TROB1 | transducer of ERBB2, 1 | Affinity Capture-Western | BioGRID | 18056425 |
| UPF1 | FLJ43809 | FLJ46894 | HUPF1 | KIAA0221 | NORF1 | RENT1 | pNORF1 | UPF1 regulator of nonsense transcripts homolog (yeast) | Affinity Capture-MS | BioGRID | 12723973 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA EIF4 PATHWAY | 24 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSLATION | 222 | 75 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | 84 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF PROTEINS | 518 | 242 | All SZGR 2.0 genes in this pathway |
| REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | 176 | 51 | All SZGR 2.0 genes in this pathway |
| REACTOME DEADENYLATION OF MRNA | 22 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF MRNA | 284 | 128 | All SZGR 2.0 genes in this pathway |
| REACTOME DEADENYLATION DEPENDENT MRNA DECAY | 48 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF RNA | 330 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | 84 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | 53 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | 176 | 57 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS TOP50 UP | 38 | 27 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA UP | 305 | 185 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR DN | 214 | 133 | All SZGR 2.0 genes in this pathway |
| LI AMPLIFIED IN LUNG CANCER | 178 | 108 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 8Q12 Q22 AMPLICON | 132 | 82 | All SZGR 2.0 genes in this pathway |
| PARK HSC VS MULTIPOTENT PROGENITORS UP | 19 | 14 | All SZGR 2.0 genes in this pathway |
| NADLER OBESITY UP | 61 | 34 | All SZGR 2.0 genes in this pathway |
| JIANG AGING CEREBRAL CORTEX UP | 36 | 27 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 24 HR DN | 91 | 64 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL DN | 128 | 93 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| LEE METASTASIS AND RNA PROCESSING UP | 17 | 9 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C D UP | 139 | 95 | All SZGR 2.0 genes in this pathway |
| CHAUHAN RESPONSE TO METHOXYESTRADIOL DN | 102 | 65 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| SWEET KRAS ONCOGENIC SIGNATURE | 89 | 56 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| WONG EMBRYONIC STEM CELL CORE | 335 | 193 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS UP | 221 | 135 | All SZGR 2.0 genes in this pathway |
| ZEMBUTSU SENSITIVITY TO NIMUSTINE | 17 | 13 | All SZGR 2.0 genes in this pathway |
| ZEMBUTSU SENSITIVITY TO VINBLASTINE | 18 | 12 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| ABRAMSON INTERACT WITH AIRE | 45 | 33 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL UP | 289 | 184 | All SZGR 2.0 genes in this pathway |