Gene Page: AKAP8L
Summary ?
| GeneID | 26993 |
| Symbol | AKAP8L |
| Synonyms | HA95|HAP95|NAKAP|NAKAP95 |
| Description | A-kinase anchoring protein 8 like |
| Reference | MIM:609475|HGNC:HGNC:29857|Ensembl:ENSG00000011243|HPRD:10641|Vega:OTTHUMG00000182446 |
| Gene type | protein-coding |
| Map location | 19p13.12 |
| Pascal p-value | 0.956 |
| Sherlock p-value | 0.877 |
| Fetal beta | 0.311 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg10403048 | 19 | 15490832 | AKAP8L | 1.96E-8 | -0.016 | 6.87E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10520921 | chr5 | 25517375 | AKAP8L | 26993 | 0.2 | trans |
Section II. Transcriptome annotation
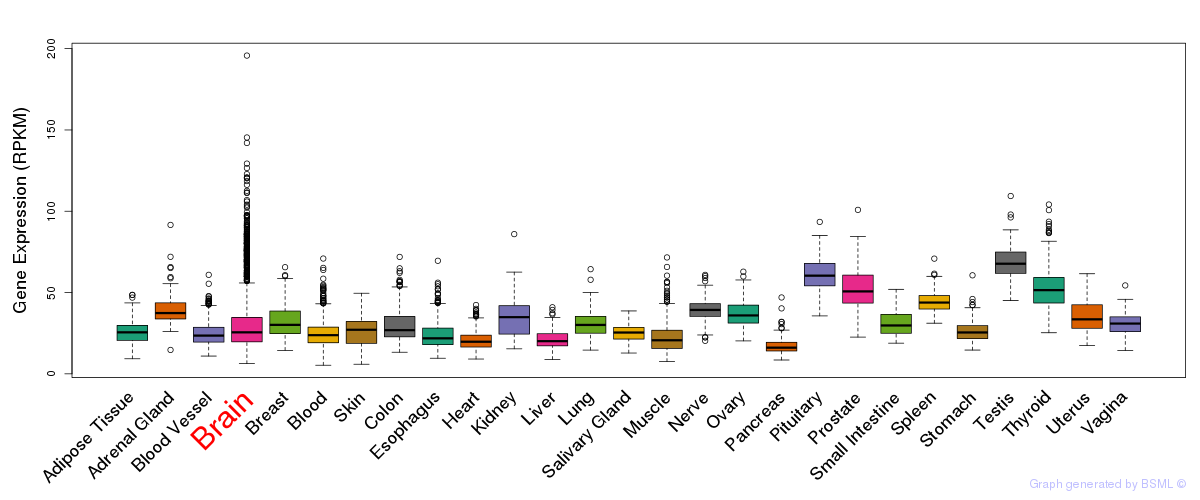
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ARL8B | 0.92 | 0.89 |
| COQ10B | 0.92 | 0.91 |
| PCMT1 | 0.91 | 0.90 |
| NAPG | 0.91 | 0.89 |
| ATP6V1B2 | 0.91 | 0.89 |
| IDH3A | 0.90 | 0.89 |
| MAP2K4 | 0.90 | 0.89 |
| MAPK9 | 0.90 | 0.87 |
| SUCLA2 | 0.90 | 0.90 |
| GLRB | 0.90 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.18 | -0.45 | -0.29 |
| IL32 | -0.44 | -0.32 |
| AF347015.21 | -0.43 | -0.23 |
| AL022328.1 | -0.41 | -0.42 |
| CLEC3B | -0.41 | -0.46 |
| AC010300.1 | -0.41 | -0.38 |
| AC098691.2 | -0.40 | -0.32 |
| AF347015.2 | -0.39 | -0.22 |
| EFEMP2 | -0.39 | -0.40 |
| AC135724.1 | -0.39 | -0.47 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| THUM SYSTOLIC HEART FAILURE DN | 244 | 147 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| YAO HOXA10 TARGETS VIA PROGESTERONE UP | 79 | 58 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR UP | 240 | 152 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 16HR UP | 225 | 139 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 8HR UP | 105 | 73 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| MAHAJAN RESPONSE TO IL1A UP | 81 | 52 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 1 | 68 | 50 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |