Gene Page: RNF11
Summary ?
| GeneID | 26994 |
| Symbol | RNF11 |
| Synonyms | CGI-123|SID1669 |
| Description | ring finger protein 11 |
| Reference | MIM:612598|HGNC:HGNC:10056|Ensembl:ENSG00000123091|HPRD:09955|Vega:OTTHUMG00000008190 |
| Gene type | protein-coding |
| Map location | 1p32 |
| Pascal p-value | 0.219 |
| Fetal beta | -1.177 |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0972 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1146358 | chr1 | 116657767 | RNF11 | 26994 | 0.17 | trans | ||
| rs1620334 | chr1 | 205697844 | RNF11 | 26994 | 0.03 | trans | ||
| rs823123 | chr1 | 205725345 | RNF11 | 26994 | 0.04 | trans | ||
| rs823066 | chr1 | 205771172 | RNF11 | 26994 | 0.02 | trans | ||
| rs11120206 | chr1 | 206610593 | RNF11 | 26994 | 0.09 | trans | ||
| rs17012898 | chr2 | 33601648 | RNF11 | 26994 | 0.03 | trans | ||
| rs17012916 | chr2 | 33606031 | RNF11 | 26994 | 0.04 | trans | ||
| rs17012924 | chr2 | 33612474 | RNF11 | 26994 | 0.08 | trans | ||
| rs2716734 | chr2 | 39947720 | RNF11 | 26994 | 9.419E-4 | trans | ||
| rs2716736 | chr2 | 39947946 | RNF11 | 26994 | 9.419E-4 | trans | ||
| rs10928499 | chr2 | 135332354 | RNF11 | 26994 | 0.06 | trans | ||
| rs17044826 | chr3 | 66620522 | RNF11 | 26994 | 0.05 | trans | ||
| rs16850133 | chr3 | 140030776 | RNF11 | 26994 | 0.13 | trans | ||
| rs17023105 | chr4 | 96074164 | RNF11 | 26994 | 1.101E-4 | trans | ||
| rs10516959 | chr4 | 96088848 | RNF11 | 26994 | 2.69E-4 | trans | ||
| rs16869878 | chr5 | 2523760 | RNF11 | 26994 | 0 | trans | ||
| rs467856 | chr5 | 3128168 | RNF11 | 26994 | 0 | trans | ||
| rs6451640 | chr5 | 20088127 | RNF11 | 26994 | 0.11 | trans | ||
| rs17103639 | chr5 | 145081858 | RNF11 | 26994 | 0.18 | trans | ||
| snp_a-2169623 | 0 | RNF11 | 26994 | 0 | trans | |||
| rs13175086 | chr5 | 174691428 | RNF11 | 26994 | 0.07 | trans | ||
| rs2016128 | chr6 | 42442743 | RNF11 | 26994 | 0 | trans | ||
| rs9376870 | chr6 | 145206590 | RNF11 | 26994 | 0.09 | trans | ||
| rs4947631 | chr7 | 50603378 | RNF11 | 26994 | 0.01 | trans | ||
| rs870686 | chr8 | 25253521 | RNF11 | 26994 | 0.03 | trans | ||
| rs10503771 | chr8 | 25304019 | RNF11 | 26994 | 0.02 | trans | ||
| rs2513775 | chr8 | 95111616 | RNF11 | 26994 | 0.15 | trans | ||
| rs12352894 | chr9 | 38197383 | RNF11 | 26994 | 0.17 | trans | ||
| rs6559219 | 0 | RNF11 | 26994 | 0.06 | trans | |||
| rs12255258 | chr10 | 106306927 | RNF11 | 26994 | 5.656E-4 | trans | ||
| rs4944475 | chr11 | 83985410 | RNF11 | 26994 | 0.01 | trans | ||
| rs7298298 | chr12 | 17810638 | RNF11 | 26994 | 0 | trans | ||
| rs998923 | chr13 | 35507810 | RNF11 | 26994 | 0.19 | trans | ||
| rs4943304 | chr13 | 36058065 | RNF11 | 26994 | 0.04 | trans | ||
| rs2518660 | chr14 | 31271351 | RNF11 | 26994 | 0.17 | trans | ||
| rs4609787 | chr15 | 61796043 | RNF11 | 26994 | 0.09 | trans | ||
| rs10512600 | chr17 | 72812978 | RNF11 | 26994 | 0.02 | trans | ||
| rs17078828 | chr18 | 65948765 | RNF11 | 26994 | 0.05 | trans | ||
| rs6040607 | chr20 | 11371092 | RNF11 | 26994 | 0.13 | trans | ||
| rs225434 | chr21 | 43724740 | RNF11 | 26994 | 6.467E-4 | trans | ||
| rs362166 | chr22 | 35461382 | RNF11 | 26994 | 0.18 | trans | ||
| rs1882710 | chrX | 149894355 | RNF11 | 26994 | 0.19 | trans |
Section II. Transcriptome annotation
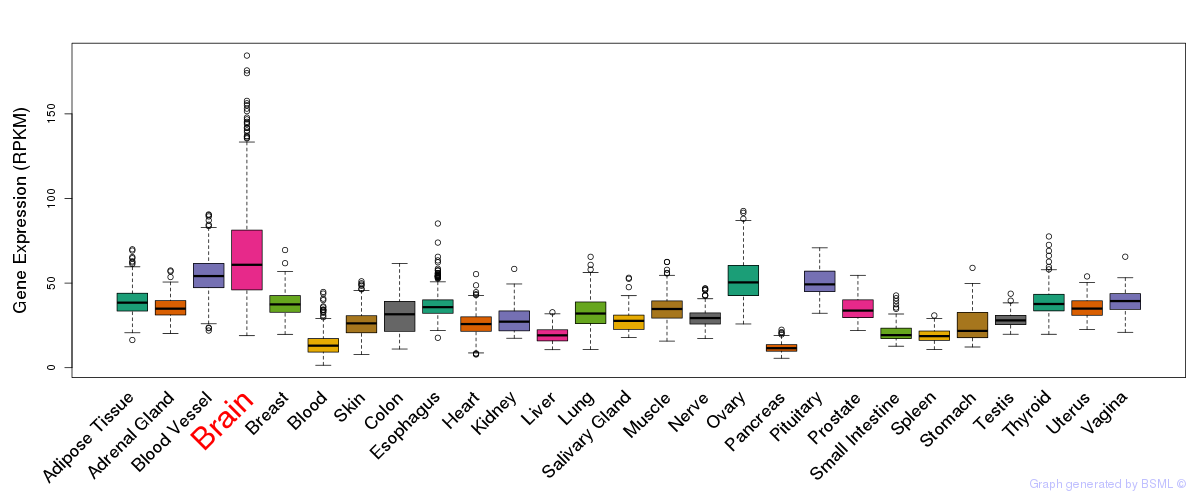
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TAF7 | 0.94 | 0.93 |
| COPS3 | 0.94 | 0.94 |
| GTF2B | 0.94 | 0.93 |
| PSMD14 | 0.94 | 0.92 |
| CCT4 | 0.94 | 0.95 |
| MRPL44 | 0.93 | 0.93 |
| ARF4 | 0.93 | 0.92 |
| CCT2 | 0.93 | 0.94 |
| CCT8 | 0.92 | 0.91 |
| MRPL3 | 0.92 | 0.92 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.33 | -0.72 | -0.80 |
| MT-CO2 | -0.72 | -0.77 |
| AF347015.8 | -0.71 | -0.79 |
| MT-CYB | -0.71 | -0.79 |
| AF347015.26 | -0.71 | -0.82 |
| AF347015.15 | -0.71 | -0.80 |
| AF347015.2 | -0.70 | -0.79 |
| AF347015.27 | -0.70 | -0.77 |
| AF347015.31 | -0.68 | -0.74 |
| HLA-F | -0.68 | -0.75 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003677 | DNA binding | TAS | 10673045 | |
| GO:0005515 | protein binding | IDA | 14755250 | |
| GO:0008270 | zinc ion binding | TAS | 10673045 | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0042787 | protein ubiquitination during ubiquitin-dependent protein catabolic process | IDA | 14755250 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000151 | ubiquitin ligase complex | IDA | 14755250 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| EPS15 | AF-1P | AF1P | MLLT5 | epidermal growth factor receptor pathway substrate 15 | Two-hybrid | BioGRID | 14755250 |
| EPS15 | AF-1P | AF1P | MLLT5 | epidermal growth factor receptor pathway substrate 15 | RNF11 interacts with EPS15. | BIND | 14755250 |
| ITCH | AIF4 | AIP4 | NAPP1 | dJ468O1.1 | itchy E3 ubiquitin protein ligase homolog (mouse) | RNF11 interacts with AIP4. | BIND | 14559117 |
| RPS27A | CEP80 | HUBCEP80 | UBA80 | UBCEP1 | UBCEP80 | ribosomal protein S27a | RNF11 interacts with RPS27A. | BIND | 14755250 |
| RPS27A | CEP80 | HUBCEP80 | UBA80 | UBCEP1 | UBCEP80 | ribosomal protein S27a | Two-hybrid | BioGRID | 14755250 |
| SMURF2 | DKFZp686F0270 | MGC138150 | SMAD specific E3 ubiquitin protein ligase 2 | RNF11 interacts with Smurf2. | BIND | 14562029 |
| STAMBP | AMSH | MGC126516 | MGC126518 | STAM binding protein | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 14755250 |
| STAMBP | AMSH | MGC126516 | MGC126518 | STAM binding protein | RNF11 interacts with AMSH. | BIND | 14755250 |
| STAMBPL1 | ALMalpha | AMSH-FP | AMSH-LP | FLJ31524 | KIAA1373 | bA399O19.2 | STAM binding protein-like 1 | Two-hybrid | BioGRID | 14755250 |
| UBC | HMG20 | ubiquitin C | RNF11 interacts with UbC. | BIND | 14755250 |
| UBC | HMG20 | ubiquitin C | Two-hybrid | BioGRID | 14755250 |
| UBE2D1 | E2(17)KB1 | SFT | UBC4/5 | UBCH5 | UBCH5A | ubiquitin-conjugating enzyme E2D 1 (UBC4/5 homolog, yeast) | RNF11 interacts with UbcH5a. | BIND | 14562029 |
| UBE2D2 | E2(17)KB2 | PUBC1 | UBC4 | UBC4/5 | UBCH5B | ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) | RNF11 interacts with UbcH5b. | BIND | 14562029 |
| UBE2D3 | E2(17)KB3 | MGC43926 | MGC5416 | UBC4/5 | UBCH5C | ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homolog, yeast) | RNF11 interacts with UbcH5c. | BIND | 14562029 |
| ZNF350 | ZBRK1 | ZFQR | zinc finger protein 350 | RNF11 interacts with ZBRK1. | BIND | 14755250 |
| ZNF350 | ZBRK1 | ZFQR | zinc finger protein 350 | Two-hybrid | BioGRID | 14755250 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PICCALUGA ANGIOIMMUNOBLASTIC LYMPHOMA DN | 136 | 86 | All SZGR 2.0 genes in this pathway |
| NAM FXYD5 TARGETS DN | 18 | 11 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS UP | 332 | 228 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA UP | 177 | 110 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS DN | 310 | 188 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS LUMINAL | 326 | 213 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR UP | 176 | 115 | All SZGR 2.0 genes in this pathway |
| GOLDRATH HOMEOSTATIC PROLIFERATION | 171 | 102 | All SZGR 2.0 genes in this pathway |
| NEMETH INFLAMMATORY RESPONSE LPS UP | 88 | 64 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION | 405 | 264 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| RUAN RESPONSE TO TNF TROGLITAZONE DN | 42 | 24 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| RUAN RESPONSE TO TNF DN | 84 | 50 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 6 | 189 | 112 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 5 | 482 | 296 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| AMUNDSON POOR SURVIVAL AFTER GAMMA RADIATION 8G | 95 | 62 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| BASSO HAIRY CELL LEUKEMIA DN | 80 | 66 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS DN | 210 | 128 | All SZGR 2.0 genes in this pathway |
| RAGHAVACHARI PLATELET SPECIFIC GENES | 70 | 46 | All SZGR 2.0 genes in this pathway |
| KARLSSON TGFB1 TARGETS DN | 207 | 139 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124.1 | 526 | 532 | m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-124/506 | 526 | 532 | 1A | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-129-5p | 465 | 471 | 1A | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-130/301 | 682 | 688 | 1A | hsa-miR-130abrain | CAGUGCAAUGUUAAAAGGGCAU |
| hsa-miR-301 | CAGUGCAAUAGUAUUGUCAAAGC | ||||
| hsa-miR-130bbrain | CAGUGCAAUGAUGAAAGGGCAU | ||||
| hsa-miR-454-3p | UAGUGCAAUAUUGCUUAUAGGGUUU | ||||
| miR-141/200a | 280 | 286 | 1A | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-186 | 393 | 399 | m8 | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
| miR-19 | 487 | 494 | 1A,m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA | ||||
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-199 | 452 | 458 | m8 | hsa-miR-199a | CCCAGUGUUCAGACUACCUGUUC |
| hsa-miR-199b | CCCAGUGUUUAGACUAUCUGUUC | ||||
| miR-24 | 62 | 68 | m8 | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
| miR-362 | 997 | 1003 | 1A | hsa-miR-362 | AAUCCUUGGAACCUAGGUGUGAGU |
| miR-369-3p | 549 | 555 | m8 | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-381 | 1170 | 1176 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-384 | 326 | 332 | 1A | hsa-miR-384 | AUUCCUAGAAAUUGUUCAUA |
| hsa-miR-384 | AUUCCUAGAAAUUGUUCAUA | ||||
| miR-496 | 200 | 206 | 1A | hsa-miR-496 | AUUACAUGGCCAAUCUC |
| miR-544 | 639 | 645 | m8 | hsa-miR-544 | AUUCUGCAUUUUUAGCAAGU |
| miR-9 | 68 | 74 | 1A | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.