Gene Page: FOXP1
Summary ?
| GeneID | 27086 |
| Symbol | FOXP1 |
| Synonyms | 12CC4|HSPC215|MFH|QRF1|hFKH1B |
| Description | forkhead box P1 |
| Reference | MIM:605515|HGNC:HGNC:3823|Ensembl:ENSG00000114861|HPRD:18518|Vega:OTTHUMG00000158803 |
| Gene type | protein-coding |
| Map location | 3p14.1 |
| Pascal p-value | 5.045E-9 |
| Sherlock p-value | 0.836 |
| Fetal beta | 0.06 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.04047 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.04359 | |
| Expression | Meta-analysis of gene expression | P value: 1.87 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg24980458 | 3 | 71632677 | FOXP1 | -0.02 | 0.41 | DMG:Nishioka_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2033050 | chr7 | 134933767 | FOXP1 | 27086 | 0.19 | trans |
Section II. Transcriptome annotation
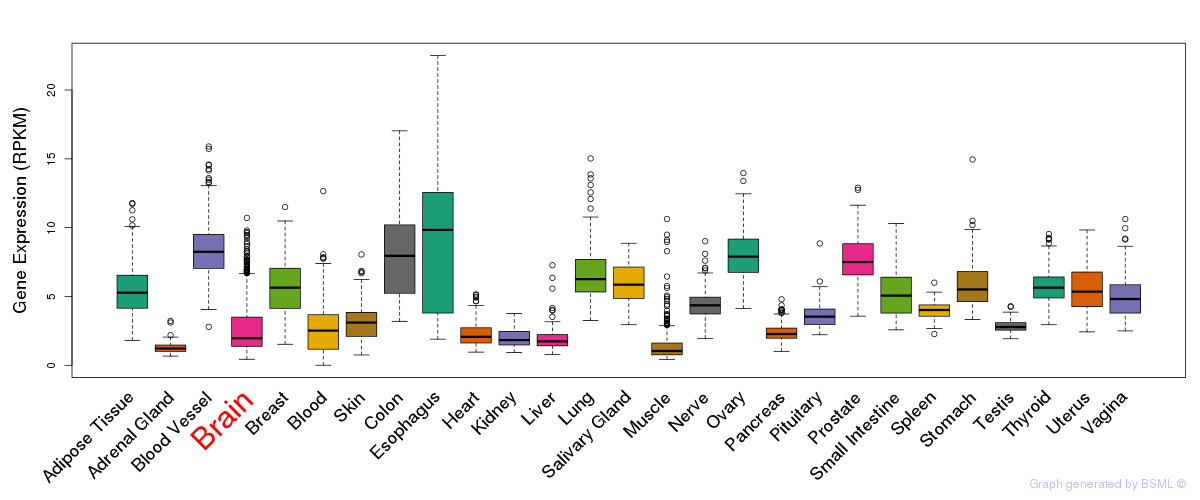
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MRTO4 | 0.79 | 0.79 |
| NARFL | 0.78 | 0.78 |
| CDK10 | 0.78 | 0.79 |
| NCAPH2 | 0.77 | 0.80 |
| RAB24 | 0.77 | 0.77 |
| PICK1 | 0.77 | 0.76 |
| TTLL3 | 0.77 | 0.77 |
| STK25 | 0.76 | 0.76 |
| RING1 | 0.76 | 0.77 |
| PNKP | 0.76 | 0.75 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.2 | -0.64 | -0.59 |
| AF347015.8 | -0.63 | -0.58 |
| AF347015.15 | -0.62 | -0.59 |
| AF347015.31 | -0.62 | -0.58 |
| MT-CYB | -0.62 | -0.59 |
| AF347015.27 | -0.61 | -0.59 |
| MT-ATP8 | -0.61 | -0.63 |
| AF347015.26 | -0.60 | -0.59 |
| MT-CO2 | -0.60 | -0.54 |
| AF347015.33 | -0.59 | -0.56 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | IEA | - | |
| GO:0003705 | RNA polymerase II transcription factor activity, enhancer binding | IEA | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0016566 | specific transcriptional repressor activity | IEA | - | |
| GO:0042803 | protein homodimerization activity | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| GO:0046982 | protein heterodimerization activity | IEA | - | |
| GO:0043565 | sequence-specific DNA binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007519 | skeletal muscle development | IEA | neuron (GO term level: 8) | - |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | IEA | - | |
| GO:0002639 | positive regulation of immunoglobulin production | IEA | - | |
| GO:0002053 | positive regulation of mesenchymal cell proliferation | IEA | - | |
| GO:0002329 | pre-B cell differentiation | IEA | - | |
| GO:0055007 | cardiac muscle cell differentiation | IEA | - | |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0009790 | embryonic development | IEA | - | |
| GO:0007507 | heart development | IEA | - | |
| GO:0016481 | negative regulation of transcription | IEA | - | |
| GO:0033152 | immunoglobulin V(D)J recombination | IEA | - | |
| GO:0030324 | lung development | IEA | - | |
| GO:0048745 | smooth muscle development | IEA | - | |
| GO:0050679 | positive regulation of epithelial cell proliferation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005730 | nucleolus | IDA | 18029348 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PICCALUGA ANGIOIMMUNOBLASTIC LYMPHOMA DN | 136 | 86 | All SZGR 2.0 genes in this pathway |
| RODRIGUES DCC TARGETS DN | 121 | 84 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS CDC25 DN | 51 | 35 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS DN | 142 | 95 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 CHRONIC LOF UP | 115 | 78 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL TGFB1 TARGETS UP | 169 | 127 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN | 514 | 319 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN | 411 | 249 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION UP | 195 | 138 | All SZGR 2.0 genes in this pathway |
| MCCLUNG CREB1 TARGETS DN | 57 | 39 | All SZGR 2.0 genes in this pathway |
| HAN JNK SINGALING UP | 35 | 21 | All SZGR 2.0 genes in this pathway |
| XU GH1 AUTOCRINE TARGETS DN | 142 | 94 | All SZGR 2.0 genes in this pathway |
| SAFFORD T LYMPHOCYTE ANERGY | 87 | 54 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 5 | 482 | 296 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| ZHENG FOXP3 TARGETS IN T LYMPHOCYTE DN | 37 | 29 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN | 409 | 268 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| COLINA TARGETS OF 4EBP1 AND 4EBP2 | 356 | 214 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN DEDIFFERENTIATED STATE DN | 7 | 7 | All SZGR 2.0 genes in this pathway |
| MARSON FOXP3 TARGETS DN | 54 | 38 | All SZGR 2.0 genes in this pathway |
| ASGHARZADEH NEUROBLASTOMA POOR SURVIVAL DN | 46 | 30 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 0 | 76 | 54 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS DN | 315 | 215 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL UP | 289 | 184 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
| ZWANG EGF PERSISTENTLY DN | 61 | 36 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-1/206 | 773 | 779 | 1A | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA | ||||
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA | ||||
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-132/212 | 2697 | 2703 | 1A | hsa-miR-212SZ | UAACAGUCUCCAGUCACGGCC |
| hsa-miR-132brain | UAACAGUCUACAGCCAUGGUCG | ||||
| miR-139 | 2808 | 2814 | m8 | hsa-miR-139brain | UCUACAGUGCACGUGUCU |
| miR-141/200a | 282 | 288 | m8 | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-150 | 172 | 178 | m8 | hsa-miR-150 | UCUCCCAACCCUUGUACCAGUG |
| miR-181 | 705 | 711 | m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-183 | 153 | 159 | m8 | hsa-miR-183 | UAUGGCACUGGUAGAAUUCACUG |
| miR-19 | 439 | 446 | 1A,m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-191 | 823 | 829 | m8 | hsa-miR-191brain | CAACGGAAUCCCAAAAGCAGCU |
| miR-22 | 2523 | 2529 | m8 | hsa-miR-22brain | AAGCUGCCAGUUGAAGAACUGU |
| miR-221/222 | 2810 | 2816 | 1A | hsa-miR-221brain | AGCUACAUUGUCUGCUGGGUUUC |
| hsa-miR-222brain | AGCUACAUCUGGCUACUGGGUCUC | ||||
| miR-30-3p | 2886 | 2893 | 1A,m8 | hsa-miR-30a-3p | CUUUCAGUCGGAUGUUUGCAGC |
| hsa-miR-30e-3p | CUUUCAGUCGGAUGUUUACAGC | ||||
| miR-324-3p | 71 | 77 | 1A | hsa-miR-324-3p | CCACUGCCCCAGGUGCUGCUGG |
| miR-329 | 540 | 546 | 1A | hsa-miR-329brain | AACACACCUGGUUAACCUCUUU |
| miR-33 | 1356 | 1362 | 1A | hsa-miR-33 | GUGCAUUGUAGUUGCAUUG |
| hsa-miR-33b | GUGCAUUGCUGUUGCAUUGCA | ||||
| miR-34/449 | 465 | 471 | m8 | hsa-miR-34abrain | UGGCAGUGUCUUAGCUGGUUGUU |
| hsa-miR-34c | AGGCAGUGUAGUUAGCUGAUUGC | ||||
| hsa-miR-449 | UGGCAGUGUAUUGUUAGCUGGU | ||||
| hsa-miR-449b | AGGCAGUGUAUUGUUAGCUGGC | ||||
| hsa-miR-34abrain | UGGCAGUGUCUUAGCUGGUUGUU | ||||
| hsa-miR-34c | AGGCAGUGUAGUUAGCUGAUUGC | ||||
| hsa-miR-449 | UGGCAGUGUAUUGUUAGCUGGU | ||||
| hsa-miR-449b | AGGCAGUGUAUUGUUAGCUGGC | ||||
| miR-34b | 466 | 472 | m8 | hsa-miR-34b | UAGGCAGUGUCAUUAGCUGAUUG |
| hsa-miR-34b | UAGGCAGUGUCAUUAGCUGAUUG | ||||
| miR-369-3p | 725 | 731 | 1A | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-374 | 725 | 731 | m8 | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-409-3p | 819 | 825 | m8 | hsa-miR-409-3p | CGAAUGUUGCUCGGUGAACCCCU |
| miR-543 | 706 | 713 | 1A,m8 | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
| miR-9 | 1730 | 1737 | 1A,m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
| hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.