Gene Page: IL36B
Summary ?
| GeneID | 27177 |
| Symbol | IL36B |
| Synonyms | FIL1|FIL1-(ETA)|FIL1H|FILI-(ETA)|IL-1F8|IL-1H2|IL1-ETA|IL1F8|IL1H2 |
| Description | interleukin 36, beta |
| Reference | MIM:605508|HGNC:HGNC:15564|Ensembl:ENSG00000136696|HPRD:05694|Vega:OTTHUMG00000131338 |
| Gene type | protein-coding |
| Map location | 2q14 |
| Pascal p-value | 0.748 |
| Fetal beta | -0.061 |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| IL36B | chr2 | 113788640 | G | A | NM_014438 NM_173178 | p.36R>C p.36R>C | missense missense | Schizophrenia | DNM:Fromer_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7370564 | 0 | IL1F8 | 27177 | 0.12 | trans |
Section II. Transcriptome annotation
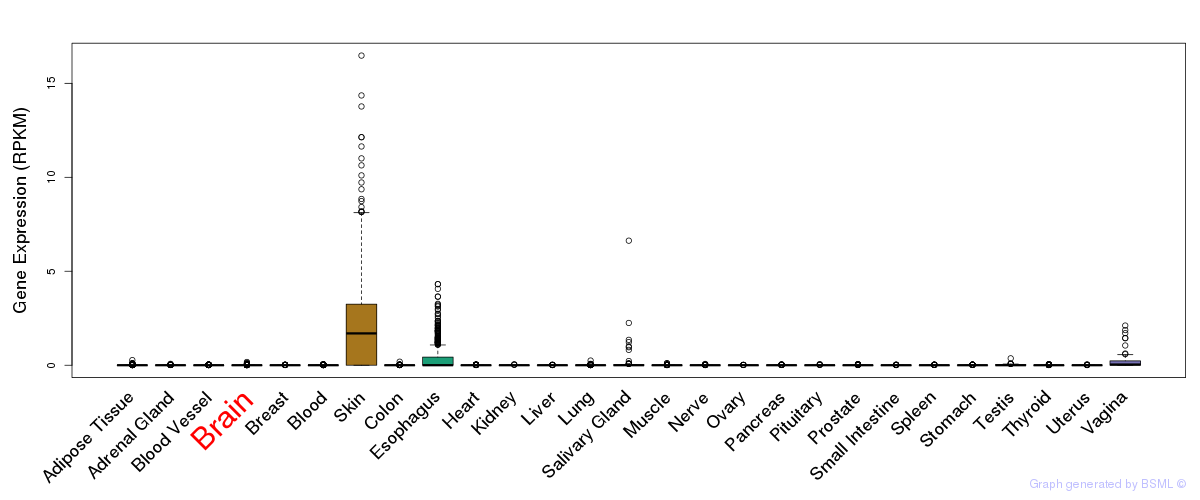
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ACD | 0.94 | 0.94 |
| ZNF446 | 0.93 | 0.93 |
| C7orf27 | 0.93 | 0.90 |
| USP21 | 0.93 | 0.91 |
| TELO2 | 0.93 | 0.92 |
| MED16 | 0.92 | 0.91 |
| GTPBP3 | 0.92 | 0.94 |
| ZNF574 | 0.92 | 0.91 |
| PTOV1 | 0.92 | 0.92 |
| ZNF408 | 0.92 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.27 | -0.73 | -0.84 |
| AF347015.31 | -0.71 | -0.82 |
| MT-CO2 | -0.71 | -0.82 |
| AF347015.33 | -0.70 | -0.80 |
| AF347015.8 | -0.70 | -0.84 |
| MT-CYB | -0.69 | -0.82 |
| AF347015.15 | -0.67 | -0.80 |
| C5orf53 | -0.65 | -0.69 |
| AF347015.2 | -0.65 | -0.80 |
| AF347015.21 | -0.64 | -0.85 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005149 | interleukin-1 receptor binding | NAS | 10744718 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006955 | immune response | NAS | 10744718 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | TAS | 10744718 | |
| GO:0005615 | extracellular space | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| HENDRICKS SMARCA4 TARGETS DN | 53 | 34 | All SZGR 2.0 genes in this pathway |
| MATTHEWS SKIN CARCINOGENESIS VIA JUN | 17 | 10 | All SZGR 2.0 genes in this pathway |
| GENTLES LEUKEMIC STEM CELL DN | 19 | 10 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |