Gene Page: AMPD3
Summary ?
| GeneID | 272 |
| Symbol | AMPD3 |
| Synonyms | - |
| Description | adenosine monophosphate deaminase 3 |
| Reference | MIM:102772|HGNC:HGNC:470|Ensembl:ENSG00000133805|HPRD:00041|Vega:OTTHUMG00000165682 |
| Gene type | protein-coding |
| Map location | 11p15 |
| Pascal p-value | 0.661 |
| Sherlock p-value | 0.085 |
| Fetal beta | -2.086 |
| DMG | 1 (# studies) |
| eGene | Caudate basal ganglia Nucleus accumbens basal ganglia Putamen basal ganglia Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 3 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg26489413 | 11 | 10476976 | AMPD3 | 4.81E-6 | -0.598 | 0.01 | DMG:Wockner_2014 |
| cg07329251 | 11 | 10476662 | AMPD3 | 6.03E-5 | -0.586 | 0.023 | DMG:Wockner_2014 |
| cg19132462 | 11 | 10476608 | AMPD3 | 1.019E-4 | -0.585 | 0.028 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs11042789 | chr11 | 10434408 | AMPD3 | 272 | 0.17 | cis | ||
| rs9971562 | 11 | 10464659 | AMPD3 | ENSG00000133805.11 | 3.93E-8 | 0 | 134799 | gtex_brain_putamen_basal |
| rs398075716 | 11 | 10466453 | AMPD3 | ENSG00000133805.11 | 1.836E-8 | 0 | 136593 | gtex_brain_putamen_basal |
| rs11605232 | 11 | 10468501 | AMPD3 | ENSG00000133805.11 | 7.128E-7 | 0 | 138641 | gtex_brain_putamen_basal |
| rs11604780 | 11 | 10468671 | AMPD3 | ENSG00000133805.11 | 5.752E-8 | 0 | 138811 | gtex_brain_putamen_basal |
| rs11606703 | 11 | 10468712 | AMPD3 | ENSG00000133805.11 | 3.013E-7 | 0 | 138852 | gtex_brain_putamen_basal |
| rs11602632 | 11 | 10468724 | AMPD3 | ENSG00000133805.11 | 5.994E-8 | 0 | 138864 | gtex_brain_putamen_basal |
| rs11604833 | 11 | 10468906 | AMPD3 | ENSG00000133805.11 | 5.987E-8 | 0 | 139046 | gtex_brain_putamen_basal |
| rs11604838 | 11 | 10468953 | AMPD3 | ENSG00000133805.11 | 5.985E-8 | 0 | 139093 | gtex_brain_putamen_basal |
| rs7929999 | 11 | 10470440 | AMPD3 | ENSG00000133805.11 | 3.003E-7 | 0 | 140580 | gtex_brain_putamen_basal |
| rs6484290 | 11 | 10470852 | AMPD3 | ENSG00000133805.11 | 2.975E-7 | 0 | 140992 | gtex_brain_putamen_basal |
| rs7112321 | 11 | 10471218 | AMPD3 | ENSG00000133805.11 | 2.967E-7 | 0 | 141358 | gtex_brain_putamen_basal |
| rs7111970 | 11 | 10471680 | AMPD3 | ENSG00000133805.11 | 1.143E-7 | 0 | 141820 | gtex_brain_putamen_basal |
| rs11042811 | 11 | 10473889 | AMPD3 | ENSG00000133805.11 | 1.494E-7 | 0 | 144029 | gtex_brain_putamen_basal |
| rs11042812 | 11 | 10474010 | AMPD3 | ENSG00000133805.11 | 1.419E-9 | 0 | 144150 | gtex_brain_putamen_basal |
| rs200902109 | 11 | 10474496 | AMPD3 | ENSG00000133805.11 | 3.765E-7 | 0 | 144636 | gtex_brain_putamen_basal |
| rs61264398 | 11 | 10474497 | AMPD3 | ENSG00000133805.11 | 5.73E-9 | 0 | 144637 | gtex_brain_putamen_basal |
| rs2071020 | 11 | 10478228 | AMPD3 | ENSG00000133805.11 | 2.771E-7 | 0 | 148368 | gtex_brain_putamen_basal |
| rs936514 | 11 | 10481613 | AMPD3 | ENSG00000133805.11 | 3.651E-9 | 0 | 151753 | gtex_brain_putamen_basal |
| rs936513 | 11 | 10481667 | AMPD3 | ENSG00000133805.11 | 3.7E-9 | 0 | 151807 | gtex_brain_putamen_basal |
| rs10840420 | 11 | 10490415 | AMPD3 | ENSG00000133805.11 | 3.788E-9 | 0 | 160555 | gtex_brain_putamen_basal |
| rs10840422 | 11 | 10491741 | AMPD3 | ENSG00000133805.11 | 1.849E-6 | 0 | 161881 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
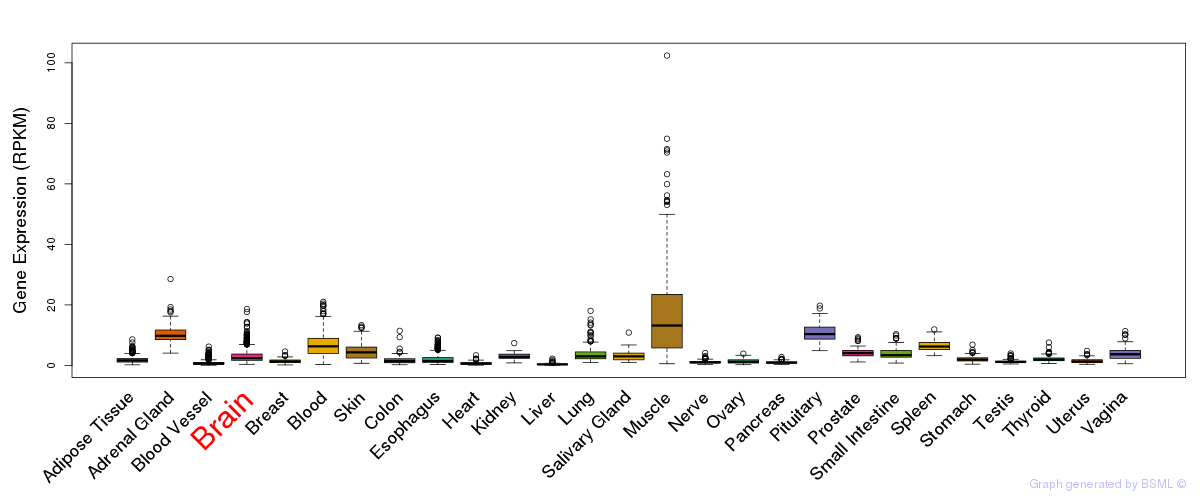
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ACADVL | 0.77 | 0.79 |
| TTC7A | 0.70 | 0.68 |
| MYO15B | 0.69 | 0.66 |
| SLC27A1 | 0.68 | 0.70 |
| PILRB | 0.67 | 0.64 |
| LAT | 0.67 | 0.61 |
| MYO15B | 0.67 | 0.66 |
| PLD2 | 0.67 | 0.73 |
| COL11A2 | 0.66 | 0.66 |
| AC010618.2 | 0.65 | 0.66 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GUCY1A2 | -0.42 | -0.42 |
| PPM1L | -0.39 | -0.33 |
| LIN7A | -0.39 | -0.39 |
| MAPK6 | -0.38 | -0.36 |
| LASS6 | -0.38 | -0.34 |
| HSPA13 | -0.37 | -0.37 |
| MOBKL3 | -0.37 | -0.40 |
| LMBRD2 | -0.37 | -0.39 |
| DOK6 | -0.37 | -0.36 |
| ZYG11B | -0.37 | -0.33 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PURINE METABOLISM | 159 | 96 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF NUCLEOTIDES | 72 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME PURINE SALVAGE | 13 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME PURINE METABOLISM | 33 | 24 | All SZGR 2.0 genes in this pathway |
| PRAMOONJAGO SOX4 TARGETS UP | 52 | 38 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 16D DN | 143 | 83 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS UP | 293 | 179 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS F DN | 33 | 24 | All SZGR 2.0 genes in this pathway |
| XU HGF TARGETS INDUCED BY AKT1 48HR UP | 14 | 8 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS DN | 261 | 155 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN | 464 | 276 | All SZGR 2.0 genes in this pathway |
| KIM GERMINAL CENTER T HELPER UP | 66 | 42 | All SZGR 2.0 genes in this pathway |
| FERNANDEZ BOUND BY MYC | 182 | 116 | All SZGR 2.0 genes in this pathway |
| LENAOUR DENDRITIC CELL MATURATION DN | 128 | 90 | All SZGR 2.0 genes in this pathway |
| MARCINIAK ER STRESS RESPONSE VIA CHOP | 25 | 17 | All SZGR 2.0 genes in this pathway |
| KIM HYPOXIA | 25 | 21 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS MATURE CELL | 293 | 160 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY DN | 145 | 88 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS UP | 388 | 234 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| GRADE COLON AND RECTAL CANCER UP | 285 | 167 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| BHATI G2M ARREST BY 2METHOXYESTRADIOL DN | 127 | 75 | All SZGR 2.0 genes in this pathway |
| ZHAN V1 LATE DIFFERENTIATION GENES UP | 32 | 25 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF LCP WITH H3K4ME3 | 128 | 68 | All SZGR 2.0 genes in this pathway |
| SETLUR PROSTATE CANCER TMPRSS2 ERG FUSION UP | 67 | 48 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC LCP WITH H3K4ME3 | 58 | 34 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 1 | 68 | 50 | All SZGR 2.0 genes in this pathway |
| HO LIVER CANCER VASCULAR INVASION | 13 | 6 | All SZGR 2.0 genes in this pathway |
| NIELSEN GIST VS SYNOVIAL SARCOMA DN | 20 | 17 | All SZGR 2.0 genes in this pathway |
| NIELSEN GIST | 98 | 66 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA MESENCHYMAL | 216 | 130 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 UP | 397 | 206 | All SZGR 2.0 genes in this pathway |
| QI HYPOXIA | 140 | 96 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS DN | 242 | 146 | All SZGR 2.0 genes in this pathway |
| ACOSTA PROLIFERATION INDEPENDENT MYC TARGETS DN | 116 | 74 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR DN | 244 | 157 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS DN | 211 | 119 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA NS UP | 162 | 103 | All SZGR 2.0 genes in this pathway |