Gene Page: LSM1
Summary ?
| GeneID | 27257 |
| Symbol | LSM1 |
| Synonyms | CASM|YJL124C |
| Description | LSM1 homolog, mRNA degradation associated |
| Reference | MIM:607281|HGNC:HGNC:20472|Ensembl:ENSG00000175324|HPRD:06281|Vega:OTTHUMG00000164051 |
| Gene type | protein-coding |
| Map location | 8p11.2 |
| Pascal p-value | 1.673E-5 |
| Sherlock p-value | 0.204 |
| Fetal beta | 0.033 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg23409476 | 8 | 38034180 | LSM1 | 1.71E-8 | -0.009 | 6.22E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
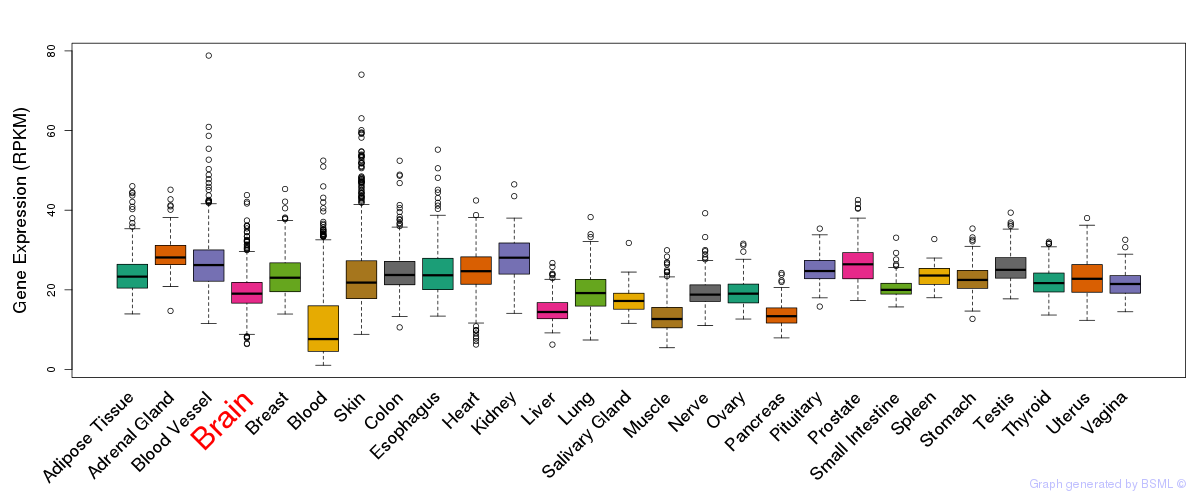
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| C3orf60 | 2P1 | DKFZp564J0123 | E3-3 | MGC10527 | chromosome 3 open reading frame 60 | Two-hybrid | BioGRID | 15231747 |
| CLIC1 | G6 | NCC27 | chloride intracellular channel 1 | Two-hybrid | BioGRID | 14667819 |
| EXOSC6 | EAP4 | MTR3 | Mtr3p | hMtr3p | p11 | exosome component 6 | Two-hybrid | BioGRID | 15231747 |
| EXOSC8 | CIP3 | EAP2 | OIP2 | RP11-421P11.3 | RRP43 | Rrp43p | bA421P11.3 | p9 | exosome component 8 | - | HPRD,BioGRID | 15231747 |
| GRHPR | GLXR | GLYD | PH2 | glyoxylate reductase/hydroxypyruvate reductase | - | HPRD,BioGRID | 15231747 |
| HSP90AA1 | FLJ31884 | HSP86 | HSP89A | HSP90A | HSP90N | HSPC1 | HSPCA | HSPCAL1 | HSPCAL4 | HSPN | Hsp89 | Hsp90 | LAP2 | heat shock protein 90kDa alpha (cytosolic), class A member 1 | - | HPRD,BioGRID | 15231747 |
| LSM2 | C6orf28 | G7b | YBL026W | snRNP | LSM2 homolog, U6 small nuclear RNA associated (S. cerevisiae) | - | HPRD,BioGRID | 11920408 |15231747 |
| LSM3 | SMX4 | USS2 | YLR438C | LSM3 homolog, U6 small nuclear RNA associated (S. cerevisiae) | - | HPRD | 11920408 |
| LSM4 | YER112W | LSM4 homolog, U6 small nuclear RNA associated (S. cerevisiae) | - | HPRD,BioGRID | 11920408 |
| LSM5 | FLJ12710 | YER146W | LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) | - | HPRD | 11920408 |
| LSM6 | YDR378C | LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) | - | HPRD | 11920408 |
| LSM7 | YNL147W | LSM7 homolog, U6 small nuclear RNA associated (S. cerevisiae) | - | HPRD | 11920408 |
| N4BP1 | FLJ31821 | KIAA0615 | MGC176730 | NEDD4 binding protein 1 | Two-hybrid | BioGRID | 15231747 |
| NARS | ASNRS | NARS1 | asparaginyl-tRNA synthetase | - | HPRD,BioGRID | 15231747 |
| PSMB5 | LMPX | MB1 | MGC104214 | X | proteasome (prosome, macropain) subunit, beta type, 5 | - | HPRD,BioGRID | 15231747 |
| PSMB8 | D6S216 | D6S216E | LMP7 | MGC1491 | PSMB5i | RING10 | beta5i | proteasome (prosome, macropain) subunit, beta type, 8 (large multifunctional peptidase 7) | - | HPRD,BioGRID | 15231747 |
| PTPRC | B220 | CD45 | CD45R | GP180 | LCA | LY5 | T200 | protein tyrosine phosphatase, receptor type, C | - | HPRD | 8980254 |
| SMN1 | BCD541 | SMA | SMA1 | SMA2 | SMA3 | SMA4 | SMA@ | SMN | SMNT | T-BCD541 | survival of motor neuron 1, telomeric | - | HPRD | 10851237 |
| TMEM214 | FLJ20254 | FLJ39682 | transmembrane protein 214 | - | HPRD,BioGRID | 15231747 |
| UPF2 | DKFZp434D222 | HUPF2 | KIAA1408 | MGC138834 | MGC138835 | RENT2 | smg-3 | UPF2 regulator of nonsense transcripts homolog (yeast) | Two-hybrid | BioGRID | 15231747 |
| UXT | ART-27 | ubiquitously-expressed transcript | - | HPRD,BioGRID | 15231747 |
| VPS11 | END1 | PEP5 | RNF108 | hVPS11 | vacuolar protein sorting 11 homolog (S. cerevisiae) | Two-hybrid | BioGRID | 15231747 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG RNA DEGRADATION | 59 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | 15 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF MRNA | 284 | 128 | All SZGR 2.0 genes in this pathway |
| REACTOME DEADENYLATION DEPENDENT MRNA DECAY | 48 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF RNA | 330 | 155 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 UP | 201 | 125 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| JAERVINEN AMPLIFIED IN LARYNGEAL CANCER | 40 | 24 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 8P12 P11 AMPLICON | 57 | 32 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER EXPRESSION BY COPY NUMBER | 100 | 62 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC | 420 | 269 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER UP | 298 | 174 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| BAE BRCA1 TARGETS UP | 75 | 47 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE EARLY LATE | 317 | 190 | All SZGR 2.0 genes in this pathway |