Gene Page: GCLM
Summary ?
| GeneID | 2730 |
| Symbol | GCLM |
| Synonyms | GLCLR |
| Description | glutamate-cysteine ligase modifier subunit |
| Reference | MIM:601176|HGNC:HGNC:4312|Ensembl:ENSG00000023909|HPRD:03106|Vega:OTTHUMG00000010562 |
| Gene type | protein-coding |
| Map location | 1p22.1 |
| Pascal p-value | 0.399 |
| Fetal beta | -0.787 |
| eGene | Cerebellum Nucleus accumbens basal ganglia Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 2 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 6 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1997108 | chr3 | 109786660 | GCLM | 2730 | 0.06 | trans | ||
| rs8071763 | chr17 | 67930673 | GCLM | 2730 | 0.01 | trans | ||
| rs871948 | chrX | 39342814 | GCLM | 2730 | 0.11 | trans |
Section II. Transcriptome annotation
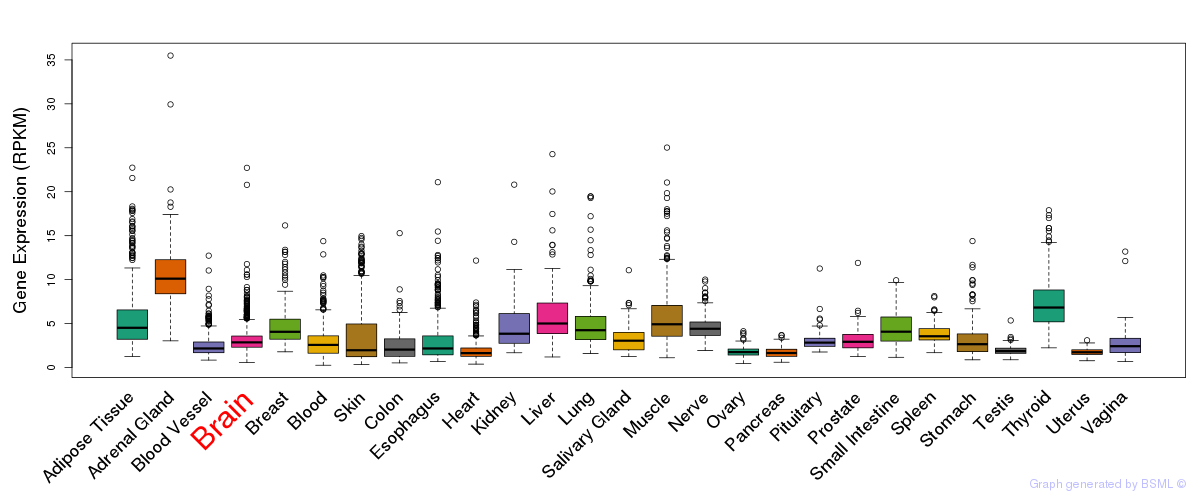
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GLUD1 | 0.95 | 0.95 |
| SUCLG2 | 0.87 | 0.85 |
| GPC5 | 0.85 | 0.85 |
| ITM2C | 0.85 | 0.85 |
| GJA1 | 0.85 | 0.83 |
| ADD3 | 0.84 | 0.88 |
| PLSCR4 | 0.83 | 0.87 |
| HADHB | 0.83 | 0.83 |
| NT5E | 0.82 | 0.84 |
| RAB31 | 0.82 | 0.81 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RTF1 | -0.54 | -0.55 |
| CCAR1 | -0.52 | -0.58 |
| RP9 | -0.52 | -0.59 |
| KIAA1949 | -0.52 | -0.39 |
| MPP3 | -0.51 | -0.46 |
| ZNF821 | -0.51 | -0.47 |
| C18orf22 | -0.51 | -0.47 |
| GMIP | -0.51 | -0.43 |
| AC011491.1 | -0.50 | -0.56 |
| DPF1 | -0.50 | -0.46 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004357 | glutamate-cysteine ligase activity | IDA | glutamate (GO term level: 6) | 9841880 |
| GO:0004357 | glutamate-cysteine ligase activity | IMP | glutamate (GO term level: 6) | 16183645 |
| GO:0035226 | glutamate-cysteine ligase catalytic subunit binding | IPI | glutamate (GO term level: 5) | 9675072 |9841880 |
| GO:0005515 | protein binding | IPI | 17353931 | |
| GO:0016491 | oxidoreductase activity | IEA | - | |
| GO:0046982 | protein heterodimerization activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006536 | glutamate metabolic process | IDA | glutamate (GO term level: 8) | 9841880 |9895302 |
| GO:0035229 | positive regulation of glutamate-cysteine ligase activity | IEA | glutamate (GO term level: 7) | - |
| GO:0006534 | cysteine metabolic process | IEA | - | |
| GO:0006750 | glutathione biosynthetic process | IDA | 10395918 | |
| GO:0006750 | glutathione biosynthetic process | IMP | 12081989 | |
| GO:0006979 | response to oxidative stress | IDA | 10395918 | |
| GO:0043066 | negative regulation of apoptosis | IEA | - | |
| GO:0042493 | response to drug | IDA | 9895302 | |
| GO:0050880 | regulation of blood vessel size | IMP | 12975258 | |
| GO:0051900 | regulation of mitochondrial depolarization | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0017109 | glutamate-cysteine ligase complex | IEA | glutamate (GO term level: 8) | - |
| GO:0005829 | cytosol | NAS | 9895302 | |
| GO:0005625 | soluble fraction | IDA | 9675072 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG GLUTATHIONE METABOLISM | 50 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME SULFUR AMINO ACID METABOLISM | 24 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | 200 | 136 | All SZGR 2.0 genes in this pathway |
| REACTOME BIOLOGICAL OXIDATIONS | 139 | 91 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUTATHIONE CONJUGATION | 23 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME PHASE II CONJUGATION | 70 | 42 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA DN | 349 | 157 | All SZGR 2.0 genes in this pathway |
| PUIFFE INVASION INHIBITED BY ASCITES UP | 82 | 51 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS YELLOW UP | 32 | 25 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS TURQUOISE UP | 79 | 50 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR DN | 214 | 133 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS UP | 293 | 179 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF DN | 228 | 137 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN UP | 239 | 157 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| DARWICHE SKIN TUMOR PROMOTER DN | 185 | 115 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK LOW UP | 162 | 104 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK HIGH UP | 147 | 101 | All SZGR 2.0 genes in this pathway |
| DARWICHE SQUAMOUS CELL CARCINOMA UP | 146 | 104 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| WOOD EBV EBNA1 TARGETS UP | 110 | 71 | All SZGR 2.0 genes in this pathway |
| LIANG HEMATOPOIESIS STEM CELL NUMBER SMALL VS HUGE UP | 38 | 29 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| HOUSTIS ROS | 36 | 29 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST DN | 309 | 206 | All SZGR 2.0 genes in this pathway |
| RAMALHO STEMNESS UP | 206 | 118 | All SZGR 2.0 genes in this pathway |
| RHODES UNDIFFERENTIATED CANCER | 69 | 44 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR UP | 156 | 101 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS DN | 215 | 132 | All SZGR 2.0 genes in this pathway |
| ZHENG RESPONSE TO ARSENITE UP | 18 | 14 | All SZGR 2.0 genes in this pathway |
| ZAMORA NOS2 TARGETS DN | 96 | 71 | All SZGR 2.0 genes in this pathway |
| GERHOLD ADIPOGENESIS UP | 49 | 40 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE UP | 393 | 244 | All SZGR 2.0 genes in this pathway |
| SINGH NFE2L2 TARGETS | 15 | 12 | All SZGR 2.0 genes in this pathway |
| BHATI G2M ARREST BY 2METHOXYESTRADIOL UP | 125 | 68 | All SZGR 2.0 genes in this pathway |
| PODAR RESPONSE TO ADAPHOSTIN UP | 147 | 98 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP | 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G1 DN | 40 | 26 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 17 | 181 | 101 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE S | 162 | 86 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
| GHANDHI BYSTANDER IRRADIATION UP | 86 | 54 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-25/32/92/363/367 | 249 | 255 | 1A | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.