Gene Page: GPX1
Summary ?
| GeneID | 2876 |
| Symbol | GPX1 |
| Synonyms | GPXD|GSHPX1 |
| Description | glutathione peroxidase 1 |
| Reference | MIM:138320|HGNC:HGNC:4553|Ensembl:ENSG00000233276|HPRD:11749|Vega:OTTHUMG00000156837 |
| Gene type | protein-coding |
| Map location | 3p21.3 |
| Pascal p-value | 0.023 |
| Fetal beta | -0.34 |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 Putamen basal ganglia |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs201181809 | 3 | 48932474 | GPX1 | ENSG00000233276.2 | 9.43071E-6 | 0.03 | 463559 | gtex_brain_ba24 |
| rs7619073 | 3 | 49125652 | GPX1 | ENSG00000233276.2 | 3.97685E-6 | 0.03 | 270381 | gtex_brain_ba24 |
| rs9883813 | 3 | 49370544 | GPX1 | ENSG00000233276.2 | 5.41001E-6 | 0.02 | 25489 | gtex_brain_putamen_basal |
| rs9854297 | 3 | 49417896 | GPX1 | ENSG00000233276.2 | 1.00928E-5 | 0.02 | -21863 | gtex_brain_putamen_basal |
| rs73073004 | 3 | 49490020 | GPX1 | ENSG00000233276.2 | 1.32611E-5 | 0.02 | -93987 | gtex_brain_putamen_basal |
| rs4855853 | 3 | 49579331 | GPX1 | ENSG00000233276.2 | 1.16399E-5 | 0.02 | -183298 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
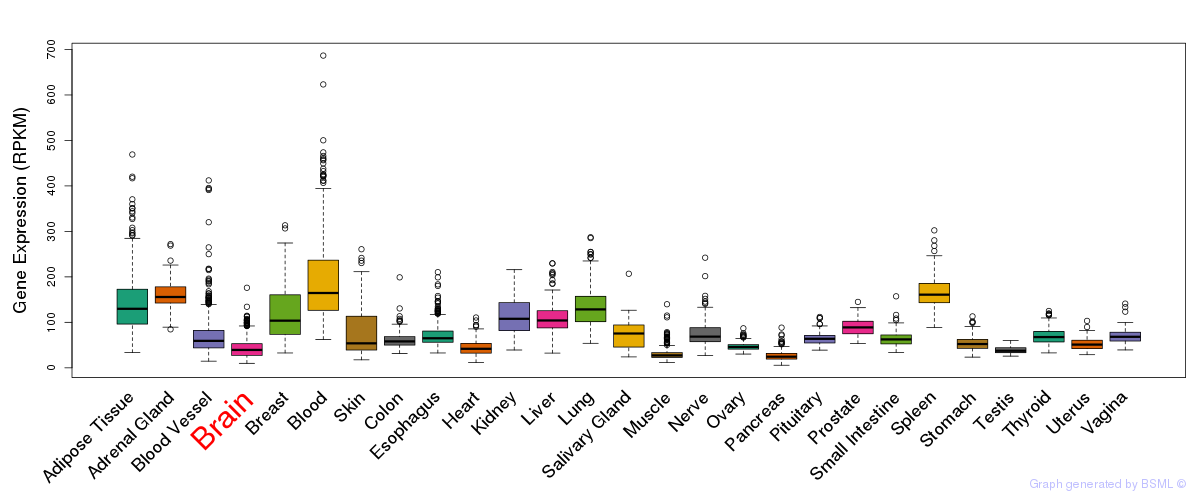
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SNX10 | 0.91 | 0.91 |
| OLFM3 | 0.90 | 0.85 |
| RAB6B | 0.89 | 0.88 |
| MOAP1 | 0.88 | 0.90 |
| MAPK9 | 0.86 | 0.92 |
| REEP5 | 0.86 | 0.87 |
| ATP1B1 | 0.86 | 0.92 |
| WBSCR17 | 0.85 | 0.88 |
| ZMAT4 | 0.85 | 0.82 |
| TPD52 | 0.85 | 0.85 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NME4 | -0.48 | -0.60 |
| SIGIRR | -0.47 | -0.56 |
| RPL28 | -0.43 | -0.51 |
| EMX2 | -0.43 | -0.57 |
| BCL7C | -0.43 | -0.52 |
| RAB13 | -0.42 | -0.53 |
| RPL23A | -0.42 | -0.45 |
| SH2D2A | -0.42 | -0.44 |
| GRTP1 | -0.41 | -0.43 |
| EFEMP2 | -0.41 | -0.39 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004602 | glutathione peroxidase activity | EXP | 8428933 | |
| GO:0004602 | glutathione peroxidase activity | IEA | - | |
| GO:0008430 | selenium binding | IEA | - | |
| GO:0008539 | proteasome inhibitor activity | IDA | 12751788 | |
| GO:0017124 | SH3 domain binding | IPI | 12893824 | |
| GO:0016491 | oxidoreductase activity | IEA | - | |
| GO:0043295 | glutathione binding | IC | 12829378 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001836 | release of cytochrome c from mitochondria | IMP | 12221075 | |
| GO:0008631 | induction of apoptosis by oxidative stress | IEA | - | |
| GO:0009650 | UV protection | IMP | 17097614 | |
| GO:0010269 | response to selenium ion | IMP | 12810669 | |
| GO:0006749 | glutathione metabolic process | IDA | 12829378 | |
| GO:0006979 | response to oxidative stress | IEA | - | |
| GO:0006916 | anti-apoptosis | IMP | 12221075 | |
| GO:0033599 | regulation of mammary gland epithelial cell proliferation | IMP | 16945136 | |
| GO:0042744 | hydrogen peroxide catabolic process | IEA | - | |
| GO:0040029 | regulation of gene expression, epigenetic | IDA | 16132718 | |
| GO:0043154 | negative regulation of caspase activity | IMP | 12221075 | |
| GO:0055114 | oxidation reduction | IEA | - | |
| GO:0060047 | heart contraction | IMP | 14573732 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 8428933 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005739 | mitochondrion | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG GLUTATHIONE METABOLISM | 50 | 34 | All SZGR 2.0 genes in this pathway |
| KEGG ARACHIDONIC ACID METABOLISM | 58 | 36 | All SZGR 2.0 genes in this pathway |
| KEGG AMYOTROPHIC LATERAL SCLEROSIS ALS | 53 | 43 | All SZGR 2.0 genes in this pathway |
| KEGG HUNTINGTONS DISEASE | 185 | 109 | All SZGR 2.0 genes in this pathway |
| BIOCARTA FREE PATHWAY | 10 | 7 | All SZGR 2.0 genes in this pathway |
| PID P53 DOWNSTREAM PATHWAY | 137 | 94 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF NUCLEOTIDES | 72 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME PURINE CATABOLISM | 10 | 6 | All SZGR 2.0 genes in this pathway |
| REACTOME PURINE METABOLISM | 33 | 24 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN | 455 | 304 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM4 | 261 | 153 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| GROSS ELK3 TARGETS UP | 27 | 16 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 UP | 209 | 139 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| HOUSTIS ROS | 36 | 29 | All SZGR 2.0 genes in this pathway |
| GOLDRATH HOMEOSTATIC PROLIFERATION | 171 | 102 | All SZGR 2.0 genes in this pathway |
| GNATENKO PLATELET SIGNATURE | 48 | 28 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| FERRANDO LYL1 NEIGHBORS | 15 | 12 | All SZGR 2.0 genes in this pathway |
| ASTIER INTEGRIN SIGNALING | 59 | 44 | All SZGR 2.0 genes in this pathway |
| TAKAO RESPONSE TO UVB RADIATION UP | 86 | 55 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR DN | 148 | 102 | All SZGR 2.0 genes in this pathway |
| ZHANG PROLIFERATING VS QUIESCENT | 51 | 41 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 24 HR DN | 91 | 64 | All SZGR 2.0 genes in this pathway |
| LEE AGING NEOCORTEX UP | 89 | 59 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL DN | 128 | 93 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| MOOTHA ROS | 7 | 7 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| ZHAN V1 LATE DIFFERENTIATION GENES UP | 32 | 25 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE UP | 181 | 106 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA MS DN | 46 | 24 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS DURATION CORR DN | 146 | 90 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS DN | 213 | 127 | All SZGR 2.0 genes in this pathway |
| HUANG GATA2 TARGETS UP | 149 | 96 | All SZGR 2.0 genes in this pathway |