Gene Page: GRB2
Summary ?
| GeneID | 2885 |
| Symbol | GRB2 |
| Synonyms | ASH|EGFRBP-GRB2|Grb3-3|MST084|MSTP084|NCKAP2 |
| Description | growth factor receptor bound protein 2 |
| Reference | MIM:108355|HGNC:HGNC:4566|Ensembl:ENSG00000177885|HPRD:00150|Vega:OTTHUMG00000134332 |
| Gene type | protein-coding |
| Map location | 17q24-q25 |
| Pascal p-value | 0.212 |
| Sherlock p-value | 0.291 |
| Fetal beta | 0.509 |
| DMG | 1 (# studies) |
| Support | INTRACELLULAR SIGNAL TRANSDUCTION G2Cdb.humanNRC G2Cdb.humanPSD G2Cdb.humanPSP Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 17.4417 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg09509576 | 17 | 73402025 | GRB2 | 2.83E-8 | -0.018 | 8.8E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
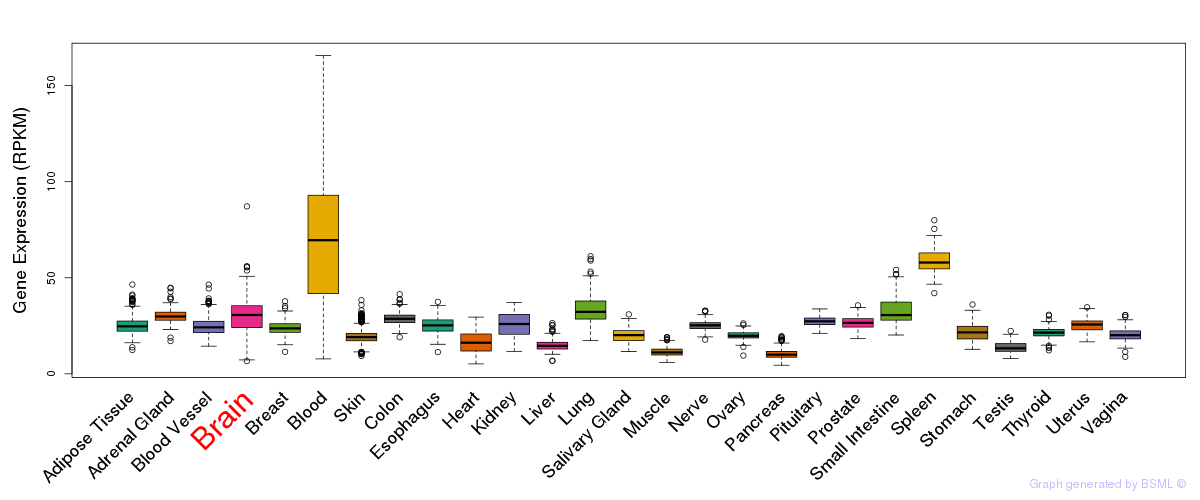
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PTPRN | 0.96 | 0.96 |
| PRRT3 | 0.94 | 0.92 |
| RASGRF1 | 0.93 | 0.89 |
| SLC12A5 | 0.93 | 0.95 |
| TRPM2 | 0.93 | 0.86 |
| KCNJ9 | 0.93 | 0.90 |
| DNM1 | 0.92 | 0.94 |
| CLSTN1 | 0.92 | 0.89 |
| CNNM1 | 0.92 | 0.92 |
| NPTN | 0.92 | 0.92 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C9orf46 | -0.47 | -0.45 |
| C21orf57 | -0.47 | -0.43 |
| BCL7C | -0.47 | -0.50 |
| RAB13 | -0.46 | -0.54 |
| RPS20 | -0.46 | -0.53 |
| GTF3C6 | -0.45 | -0.41 |
| RPS6 | -0.44 | -0.44 |
| RPL23A | -0.43 | -0.46 |
| RPS23 | -0.43 | -0.39 |
| RPL24 | -0.43 | -0.41 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005154 | epidermal growth factor receptor binding | IPI | 12577067 | |
| GO:0005070 | SH3/SH2 adaptor activity | TAS | 8253073 | |
| GO:0043560 | insulin receptor substrate binding | IPI | 8388384 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007173 | epidermal growth factor receptor signaling pathway | TAS | 1322798 | |
| GO:0007267 | cell-cell signaling | TAS | 8253073 | |
| GO:0007265 | Ras protein signal transduction | EXP | 11520933 | |
| GO:0007265 | Ras protein signal transduction | TAS | 8253073 | |
| GO:0008286 | insulin receptor signaling pathway | IPI | 8388384 | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005794 | Golgi apparatus | IEA | - | |
| GO:0005829 | cytosol | EXP | 7739560 |8479541 |8493579 |8621719 |8810325 |9356464 |9690470 |10567358 |10648629 |10734310 |10913131 |11606067 | |
| GO:0005829 | cytosol | TAS | 14722116 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABL1 | ABL | JTK7 | bcr/abl | c-ABL | p150 | v-abl | c-abl oncogene 1, receptor tyrosine kinase | - | HPRD | 9516488 |
| ABL1 | ABL | JTK7 | bcr/abl | c-ABL | p150 | v-abl | c-abl oncogene 1, receptor tyrosine kinase | Affinity Capture-Western | BioGRID | 9407116 |
| ACTA1 | ACTA | ASMA | CFTD | CFTD1 | CFTDM | MPFD | NEM1 | NEM2 | NEM3 | actin, alpha 1, skeletal muscle | Affinity Capture-MS | BioGRID | 12577067 |
| ADA | - | adenosine deaminase | - | HPRD | 9299436 |
| ADAM10 | AD10 | CD156c | HsT18717 | MADM | kuz | ADAM metallopeptidase domain 10 | Affinity Capture-Western | BioGRID | 11741929 |
| ADAM12 | MCMP | MCMPMltna | MLTN | MLTNA | ADAM metallopeptidase domain 12 | - | HPRD | 11127814 |
| ADAM15 | MDC15 | ADAM metallopeptidase domain 15 | - | HPRD,BioGRID | 11741929 |
| ADRB1 | ADRB1R | B1AR | BETA1AR | RHR | adrenergic, beta-1-, receptor | beta-1-AR interacts with Grb2. | BIND | 9843378 |
| ADRB2 | ADRB2R | ADRBR | B2AR | BAR | BETA2AR | adrenergic, beta-2-, receptor, surface | - | HPRD,BioGRID | 9830057 |
| ALOX5 | 5-LO | 5-LOX | 5LPG | LOG5 | MGC163204 | arachidonate 5-lipoxygenase | - | HPRD,BioGRID | 7929073 |8585605 |
| ANXA2 | ANX2 | ANX2L4 | CAL1H | LIP2 | LPC2 | LPC2D | P36 | PAP-IV | annexin A2 | - | HPRD,BioGRID | 7510700 |
| APP | AAA | ABETA | ABPP | AD1 | APPI | CTFgamma | CVAP | PN2 | amyloid beta (A4) precursor protein | - | HPRD | 12485888 |
| ARHGAP17 | DKFZp564A1363 | FLJ37567 | FLJ43368 | MGC87805 | MST066 | MST110 | MSTP038 | MSTP066 | MSTP110 | NADRIN | RICH1 | WBP15 | Rho GTPase activating protein 17 | Reconstituted Complex | BioGRID | 11431473 |
| AXL | JTK11 | UFO | AXL receptor tyrosine kinase | Axl interacts with an unspecified isoform of Grb2. | BIND | 12470648 |
| AXL | JTK11 | UFO | AXL receptor tyrosine kinase | - | HPRD,BioGRID | 9178760 |
| BAT2 | D6S51 | D6S51E | DKFZp686D09175 | G2 | HLA-B associated transcript 2 | - | HPRD,BioGRID | 14667819 |
| BCAR1 | CAS | CAS1 | CASS1 | CRKAS | P130Cas | breast cancer anti-estrogen resistance 1 | - | HPRD | 8649368 |8700507 |
| BCAR1 | CAS | CAS1 | CASS1 | CRKAS | P130Cas | breast cancer anti-estrogen resistance 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 10085298 |10822386 |
| BCL2A1 | ACC-1 | ACC-2 | BCL2L5 | BFL1 | GRS | HBPA1 | BCL2-related protein A1 | - | HPRD | 9670936 |
| BCR | ALL | BCR-ABL1 | BCR1 | CML | D22S11 | D22S662 | FLJ16453 | PHL | breakpoint cluster region | Grb2 interacts with Bcr-Abl. | BIND | 9872323 |
| BCR | ALL | BCR-ABL1 | BCR1 | CML | D22S11 | D22S662 | FLJ16453 | PHL | breakpoint cluster region | - | HPRD,BioGRID | 8112292 |9178913 |
| BCR | ALL | BCR-ABL1 | BCR1 | CML | D22S11 | D22S662 | FLJ16453 | PHL | breakpoint cluster region | Tyrosine phosphorylated Bcr-Abl interacts with Grb2. | BIND | 8112292 |
| BLNK | BASH | BLNK-S | LY57 | MGC111051 | SLP-65 | SLP65 | B-cell linker | - | HPRD,BioGRID | 9697839 |9705962 |
| CALD1 | CDM | H-CAD | L-CAD | MGC21352 | NAG22 | caldesmon 1 | - | HPRD | 10559276 |
| CBL | C-CBL | CBL2 | RNF55 | Cas-Br-M (murine) ecotropic retroviral transforming sequence | Grb2 interacts with Cbl. | BIND | 15782196 |
| CBL | C-CBL | CBL2 | RNF55 | Cas-Br-M (murine) ecotropic retroviral transforming sequence | c-Cbl interacts with Grb2. This interaction was modeled on a demonstrated interaction between human c-Cbl and Grb2 from an unspecified species. | BIND | 11053437 |
| CBL | C-CBL | CBL2 | RNF55 | Cas-Br-M (murine) ecotropic retroviral transforming sequence | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 8083187 |9108392 |9174058 |9178909 |9367879 |9461587 |9872323 |10086340 |10204582 |10570290 |11964172 |11997436 |
| CBL | C-CBL | CBL2 | RNF55 | Cas-Br-M (murine) ecotropic retroviral transforming sequence | - | HPRD | 9108392 |9178909 |11133830 |11964172 |
| CBLB | DKFZp686J10223 | DKFZp779A0729 | DKFZp779F1443 | FLJ36865 | FLJ41152 | Nbla00127 | RNF56 | Cas-Br-M (murine) ecotropic retroviral transforming sequence b | - | HPRD,BioGRID | 10086340 |
| CCL5 | D17S136E | MGC17164 | RANTES | SCYA5 | SISd | TCP228 | chemokine (C-C motif) ligand 5 | - | HPRD | 9009162 |
| CD22 | FLJ22814 | MGC130020 | SIGLEC-2 | SIGLEC2 | CD22 molecule | - | HPRD,BioGRID | 11551923 |
| CD28 | MGC138290 | Tp44 | CD28 molecule | - | HPRD,BioGRID | 7737275 |8576157 |9694876 |
| CD2AP | CMS | DKFZp586H0519 | CD2-associated protein | - | HPRD | 10339567 |
| CD72 | CD72b | LYB2 | CD72 molecule | - | HPRD | 10820378 |
| CDKN1B | CDKN4 | KIP1 | MEN1B | MEN4 | P27KIP1 | cyclin-dependent kinase inhibitor 1B (p27, Kip1) | - | HPRD,BioGRID | 11278754 |
| CHRM4 | HM4 | cholinergic receptor, muscarinic 4 | M4 interacts with Grb2. | BIND | 9843378 |
| CLYBL | CLB | bA134O15.1 | citrate lyase beta like | Affinity Capture-MS | BioGRID | 12577067 |
| CRK | CRKII | v-crk sarcoma virus CT10 oncogene homolog (avian) | Affinity Capture-Western Far Western Reconstituted Complex | BioGRID | 8662907 |8810325 |10970810 |
| CRK | CRKII | v-crk sarcoma virus CT10 oncogene homolog (avian) | - | HPRD | 8662907 |10970810 |
| CRKL | - | v-crk sarcoma virus CT10 oncogene homolog (avian)-like | - | HPRD,BioGRID | 10477741 |
| CSF1R | C-FMS | CD115 | CSFR | FIM2 | FMS | colony stimulating factor 1 receptor | - | HPRD,BioGRID | 8262059 |9178909 |9380408 |
| CSF3R | CD114 | GCSFR | colony stimulating factor 3 receptor (granulocyte) | - | HPRD,BioGRID | 9824671 |
| CTTN | EMS1 | FLJ34459 | cortactin | - | HPRD | 11439336 |
| DAG1 | 156DAG | A3a | AGRNR | DAG | dystroglycan 1 (dystrophin-associated glycoprotein 1) | - | HPRD,BioGRID | 7744812 |
| DCTN1 | DAP-150 | DP-150 | HMN7B | P135 | dynactin 1 (p150, glued homolog, Drosophila) | - | HPRD,BioGRID | 8955163 |
| DNM1 | DNM | dynamin 1 | Affinity Capture-Western Far Western Reconstituted Complex | BioGRID | 8119878 |9009162 |
| DNM1 | DNM | dynamin 1 | - | HPRD | 8119878 |11877424 |
| DOCK1 | DOCK180 | ced5 | dedicator of cytokinesis 1 | Co-fractionation Reconstituted Complex | BioGRID | 8657152 |12615911 |
| DRD3 | D3DR | ETM1 | FET1 | MGC149204 | MGC149205 | dopamine receptor D3 | D3 interacts with Grb2. | BIND | 9843378 |
| DRD3 | D3DR | ETM1 | FET1 | MGC149204 | MGC149205 | dopamine receptor D3 | - | HPRD,BioGRID | 11384839 |
| DRD4 | D4DR | dopamine receptor D4 | D4.7 interacts with Grb2. | BIND | 9843378 |
| DRD4 | D4DR | dopamine receptor D4 | D4.4 interacts with Grb2. | BIND | 9843378 |
| DRD4 | D4DR | dopamine receptor D4 | - | HPRD,BioGRID | 9843378 |
| DRD4 | D4DR | dopamine receptor D4 | D4.2 interacts with Grb2. | BIND | 9843378 |
| DTX1 | hDx-1 | deltex homolog 1 (Drosophila) | - | HPRD,BioGRID | 9590294 |
| EGF | HOMG4 | URG | epidermal growth factor (beta-urogastrone) | - | HPRD | 10973965 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | GRB2 interacts with an unspecified isoform of EGFR. This interaction was modeled on a demonstrated interaction between human GRB2, and rat EGFR. | BIND | 15782189 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | - | HPRD | 1322798 |7527043 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | EGFR interacts with GRB2. | BIND | 8305738 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | Affinity Capture-MS Affinity Capture-Western Reconstituted Complex | BioGRID | 1322798 |7510700 |7527043 |8647858 |9050991 |10026169 |10085134 |11960376 |12577067 |16729043 |
| ELK1 | - | ELK1, member of ETS oncogene family | - | HPRD | 8798570 |
| EPHA2 | ECK | EPH receptor A2 | - | HPRD,BioGRID | 12400011 |
| EPHB1 | ELK | EPHT2 | FLJ37986 | Hek6 | NET | EPH receptor B1 | - | HPRD,BioGRID | 8798570 |
| EPOR | MGC138358 | erythropoietin receptor | - | HPRD,BioGRID | 7534299 |
| EPPK1 | EPIPL | EPIPL1 | epiplakin 1 | Affinity Capture-MS | BioGRID | 12577067 |
| EPS15 | AF-1P | AF1P | MLLT5 | epidermal growth factor receptor pathway substrate 15 | Far Western | BioGRID | 7797522 |
| ERBB2 | CD340 | HER-2 | HER-2/neu | HER2 | NEU | NGL | TKR1 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) | - | HPRD | 11173924 |11606575|11606575 |
| ERBB2 | CD340 | HER-2 | HER-2/neu | HER2 | NEU | NGL | TKR1 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) | - | HPRD,BioGRID | 11606575 |
| ERBB3 | ErbB-3 | HER3 | LCCS2 | MDA-BF-1 | MGC88033 | c-erbB-3 | c-erbB3 | erbB3-S | p180-ErbB3 | p45-sErbB3 | p85-sErbB3 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian) | Affinity Capture-MS | BioGRID | 16729043 |
| ERBB4 | HER4 | MGC138404 | p180erbB4 | v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) | Affinity Capture-MS | BioGRID | 16729043 |
| ETV6 | TEL | TEL/ABL | ets variant 6 | Affinity Capture-Western Far Western | BioGRID | 15143164 |
| FASLG | APT1LG1 | CD178 | CD95L | FASL | TNFSF6 | Fas ligand (TNF superfamily, member 6) | - | HPRD,BioGRID | 11741599 |12023017 |
| FASLG | APT1LG1 | CD178 | CD95L | FASL | TNFSF6 | Fas ligand (TNF superfamily, member 6) | CD95L interacts with Grb2. | BIND | 12023017 |
| FGFR1 | BFGFR | CD331 | CEK | FGFBR | FLG | FLJ99988 | FLT2 | HBGFR | KAL2 | N-SAM | fibroblast growth factor receptor 1 | - | HPRD | 9480847 |
| FGFR3 | ACH | CD333 | CEK2 | HSFGFR3EX | JTK4 | fibroblast growth factor receptor 3 | - | HPRD,BioGRID | 9045692 |
| FLT3 | CD135 | FLK2 | STK1 | fms-related tyrosine kinase 3 | - | HPRD,BioGRID | 10080542 |
| FLT4 | FLT41 | LMPH1A | PCL | VEGFR3 | fms-related tyrosine kinase 4 | - | HPRD,BioGRID | 7675451 |7970715 |8662748 |9927207 |
| FRS2 | FRS2A | FRS2alpha | SNT | SNT-1 | SNT1 | fibroblast growth factor receptor substrate 2 | Affinity Capture-Western Reconstituted Complex | BioGRID | 8780727 |9182757 |10092678 |11997436 |
| FRS2 | FRS2A | FRS2alpha | SNT | SNT-1 | SNT1 | fibroblast growth factor receptor substrate 2 | - | HPRD | 8780727|9182757|11997436 |12402043 |
| FRS3 | FRS2B | FRS2beta | MGC17167 | SNT-2 | SNT2 | fibroblast growth factor receptor substrate 3 | - | HPRD | 11432792 |
| GAB1 | - | GRB2-associated binding protein 1 | - | HPRD,BioGRID | 11314042 |11960376 |
| GAB2 | KIAA0571 | GRB2-associated binding protein 2 | Affinity Capture-Western Reconstituted Complex | BioGRID | 10391903 |11782427 |15143164 |
| GAB2 | KIAA0571 | GRB2-associated binding protein 2 | - | HPRD | 10068651 |11882361 |
| GAB3 | - | GRB2-associated binding protein 3 | - | HPRD,BioGRID | 11739737 |
| GHR | GHBP | growth hormone receptor | - | HPRD,BioGRID | 9632636 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD,BioGRID | 7716522 |
| GRB7 | - | growth factor receptor-bound protein 7 | - | HPRD | 10377264 |
| GRLF1 | GRF-1 | KIAA1722 | MGC10745 | P190-A | P190A | p190RhoGAP | glucocorticoid receptor DNA binding factor 1 | Affinity Capture-Western | BioGRID | 12441060 |
| HCLS1 | CTTNL | HS1 | hematopoietic cell-specific Lyn substrate 1 | Reconstituted Complex | BioGRID | 9670936 |
| HNRNPC | C1 | C2 | HNRNP | HNRPC | MGC104306 | MGC105117 | MGC117353 | MGC131677 | SNRPC | heterogeneous nuclear ribonucleoprotein C (C1/C2) | - | HPRD,BioGRID | 9516488 |
| HRAS | C-BAS/HAS | C-H-RAS | C-HA-RAS1 | CTLO | H-RASIDX | HAMSV | HRAS1 | K-RAS | N-RAS | RASH1 | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | - | HPRD | 11352655 |
| HTT | HD | IT15 | huntingtin | - | HPRD,BioGRID | 9079622 |
| HTT | HD | IT15 | huntingtin | HDP interacts with Grb2. | BIND | 9079622 |
| IL2RB | CD122 | P70-75 | interleukin 2 receptor, beta | - | HPRD,BioGRID | 10602027 |
| IL6ST | CD130 | CDw130 | GP130 | GP130-RAPS | IL6R-beta | interleukin 6 signal transducer (gp130, oncostatin M receptor) | Affinity Capture-Western Reconstituted Complex | BioGRID | 9013873 |
| INPP5D | MGC104855 | MGC142140 | MGC142142 | SHIP | SHIP1 | SIP-145 | hp51CN | inositol polyphosphate-5-phosphatase, 145kDa | - | HPRD,BioGRID | 8723348 |
| INSR | CD220 | HHF5 | insulin receptor | Affinity Capture-Western | BioGRID | 9083103 |
| IQGAP1 | HUMORFA01 | KIAA0051 | SAR1 | p195 | IQ motif containing GTPase activating protein 1 | Affinity Capture-MS | BioGRID | 12577067 |
| IRS1 | HIRS-1 | insulin receptor substrate 1 | - | HPRD | 7488107 |
| IRS1 | HIRS-1 | insulin receptor substrate 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 8491186 |8536716 |12173038 |
| IRS2 | - | insulin receptor substrate 2 | - | HPRD | 9852124 |
| IRS4 | IRS-4 | PY160 | insulin receptor substrate 4 | - | HPRD,BioGRID | 9553137 |
| ITGA2B | CD41 | CD41B | GP2B | GPIIb | GTA | HPA3 | integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) | - | HPRD,BioGRID | 8631894 |11964172 |
| ITGA6 | CD49f | DKFZp686J01244 | FLJ18737 | ITGA6B | VLA-6 | integrin, alpha 6 | - | HPRD | 7556090 |
| ITGB4 | CD104 | integrin, beta 4 | - | HPRD | 9171350 |
| ITK | EMT | LYK | MGC126257 | MGC126258 | PSCTK2 | IL2-inducible T-cell kinase | - | HPRD,BioGRID | 10636929 |
| JAK1 | JAK1A | JAK1B | JTK3 | Janus kinase 1 (a protein tyrosine kinase) | - | HPRD,BioGRID | 8536716 |11527382 |
| JAK2 | JTK10 | Janus kinase 2 (a protein tyrosine kinase) | - | HPRD,BioGRID | 7500025 |
| JUB | Ajuba | MGC15563 | jub, ajuba homolog (Xenopus laevis) | - | HPRD | 10330178 |
| KDR | CD309 | FLK1 | VEGFR | VEGFR2 | kinase insert domain receptor (a type III receptor tyrosine kinase) | - | HPRD,BioGRID | 10319320 |
| KHDRBS1 | FLJ34027 | Sam68 | p62 | KH domain containing, RNA binding, signal transduction associated 1 | - | HPRD | 7799925 |8576157 |12112020 |
| KHDRBS1 | FLJ34027 | Sam68 | p62 | KH domain containing, RNA binding, signal transduction associated 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 9872323 |10467411 |11960376 |
| KHDRBS1 | FLJ34027 | Sam68 | p62 | KH domain containing, RNA binding, signal transduction associated 1 | Grb2 interacts with Sam68. | BIND | 9872323 |
| KHDRBS2 | FLJ38664 | MGC26664 | SLM-1 | SLM1 | bA535F17.1 | KH domain containing, RNA binding, signal transduction associated 2 | - | HPRD | 10077576 |
| KIT | C-Kit | CD117 | PBT | SCFR | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | Kit interacts with Grb2. This interaction was modeled on a demonstrated interaction between mouse proteins. | BIND | 10022833 |
| KIT | C-Kit | CD117 | PBT | SCFR | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | Grb2 interacts with c-Kit upon stimulation with SLF. This interaction was modeled on a demonstrated interaction between mouse Grb2 and human c-Kit. | BIND | 7523381 |
| KIT | C-Kit | CD117 | PBT | SCFR | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | - | HPRD,BioGRID | 10377264 |
| KRT17 | K17 | PC | PC2 | PCHC1 | keratin 17 | Affinity Capture-MS | BioGRID | 12577067 |
| KRT18 | CYK18 | K18 | keratin 18 | Affinity Capture-MS | BioGRID | 12577067 |
| KRT7 | CK7 | K2C7 | K7 | MGC129731 | MGC3625 | SCL | keratin 7 | Affinity Capture-MS | BioGRID | 12577067 |
| KRT8 | CARD2 | CK8 | CYK8 | K2C8 | K8 | KO | keratin 8 | Affinity Capture-MS | BioGRID | 12577067 |
| LAT | LAT1 | pp36 | linker for activation of T cells | - | HPRD | 9489702 |12359715 |
| LAT | LAT1 | pp36 | linker for activation of T cells | Affinity Capture-Western Reconstituted Complex | BioGRID | 9489702 |11368773 |12186560 |
| LAT2 | HSPC046 | LAB | NTAL | WBSCR15 | WBSCR5 | WSCR5 | linker for activation of T cells family, member 2 | - | HPRD,BioGRID | 12514734 |
| LAX1 | FLJ20340 | LAX | lymphocyte transmembrane adaptor 1 | - | HPRD,BioGRID | 12359715 |
| LCP2 | SLP-76 | SLP76 | lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) | Affinity Capture-Western Reconstituted Complex | BioGRID | 8695800 |8995445 |10204582 |10224278 |11314042 |
| LY6G6F | C6orf21 | G6f | LY6G6D | NG32 | lymphocyte antigen 6 complex, locus G6F | - | HPRD,BioGRID | 12852788 |
| MAP2 | DKFZp686I2148 | MAP2A | MAP2B | MAP2C | microtubule-associated protein 2 | - | HPRD,BioGRID | 10781592 |11546790|11546790 |
| MAP3K1 | MAPKKK1 | MEKK | MEKK1 | mitogen-activated protein kinase kinase kinase 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 9733714 |
| MAP4K1 | HPK1 | mitogen-activated protein kinase kinase kinase kinase 1 | - | HPRD,BioGRID | 9346925 |9788432 |9891069 |
| MAP4K1 | HPK1 | mitogen-activated protein kinase kinase kinase kinase 1 | HPK1, a Ste-20 homologous serine/threonine kinase interacts with Grb2, an adaptor protein. | BIND | 9891069 |
| MAPK12 | ERK3 | ERK6 | P38GAMMA | PRKM12 | SAPK-3 | SAPK3 | mitogen-activated protein kinase 12 | - | HPRD | 9019171 |
| MAPK14 | CSBP1 | CSBP2 | CSPB1 | EXIP | Mxi2 | PRKM14 | PRKM15 | RK | SAPK2A | p38 | p38ALPHA | mitogen-activated protein kinase 14 | - | HPRD | 8695800 |
| MAPK9 | JNK-55 | JNK2 | JNK2A | JNK2ALPHA | JNK2B | JNK2BETA | PRKM9 | SAPK | p54a | p54aSAPK | mitogen-activated protein kinase 9 | Reconstituted Complex | BioGRID | 7642542 |9019171 |
| MED28 | 1500003D12Rik | DKFZp434N185 | EG1 | magicin | mediator complex subunit 28 | - | HPRD,BioGRID | 15467741 |
| MERTK | MER | MGC133349 | RP38 | c-mer | c-mer proto-oncogene tyrosine kinase | - | HPRD | 9891051 |
| MET | AUTS9 | HGFR | RCCP2 | c-Met | met proto-oncogene (hepatocyte growth factor receptor) | - | HPRD,BioGRID | 7511210 |8662889 |9660480 |
| MICAL1 | DKFZp434B1517 | FLJ11937 | FLJ21739 | MICAL | MICAL-1 | NICAL | microtubule associated monoxygenase, calponin and LIM domain containing 1 | Reconstituted Complex | BioGRID | 11827972 |
| MST1R | CD136 | CDw136 | PTK8 | RON | macrophage stimulating 1 receptor (c-met-related tyrosine kinase) | - | HPRD,BioGRID | 8918464 |
| MTA1 | - | metastasis associated 1 | - | HPRD | 11483358 |
| MTA3 | KIAA1266 | metastasis associated 1 family, member 3 | - | HPRD | 11483358 |
| MUC1 | CD227 | EMA | H23AG | MAM6 | PEM | PEMT | PUM | mucin 1, cell surface associated | - | HPRD,BioGRID | 7664271 |
| NCKIPSD | AF3P21 | DIP | DIP1 | MGC23891 | ORF1 | SPIN90 | WASLBP | WISH | NCK interacting protein with SH3 domain | - | HPRD | 11157975 |11509578 |
| NCKIPSD | AF3P21 | DIP | DIP1 | MGC23891 | ORF1 | SPIN90 | WASLBP | WISH | NCK interacting protein with SH3 domain | - | HPRD,BioGRID | 11157975 |
| NEU3 | FLJ12388 | SIAL3 | sialidase 3 (membrane sialidase) | - | HPRD,BioGRID | 12730204 |
| NTRK1 | DKFZp781I14186 | MTC | TRK | TRK1 | TRKA | p140-TrkA | neurotrophic tyrosine kinase, receptor, type 1 | - | HPRD,BioGRID | 10748052 |11733534 |
| OCRL | INPP5F | LOCR | NPHL2 | OCRL1 | oculocerebrorenal syndrome of Lowe | - | HPRD,BioGRID | 9038219 |
| PAG1 | CBP | FLJ37858 | MGC138364 | PAG | phosphoprotein associated with glycosphingolipid microdomains 1 | - | HPRD,BioGRID | 10790433 |
| PAK1 | MGC130000 | MGC130001 | PAKalpha | p21 protein (Cdc42/Rac)-activated kinase 1 | PAK1 interacts with Grb2. | BIND | 12522133 |
| PDE4D | DPDE3 | HSPDE4D | PDE4DN2 | STRK1 | phosphodiesterase 4D, cAMP-specific (phosphodiesterase E3 dunce homolog, Drosophila) | PDE4D4 interacts with Grb2. This interaction was modelled on a demonstrated interaction between human PDE4D4 and Grb2 from an unspecified species. | BIND | 10571082 |
| PDE6G | DKFZp686C0587 | MGC125749 | PDEG | phosphodiesterase 6G, cGMP-specific, rod, gamma | - | HPRD,BioGRID | 12624098 |
| PDGFRA | CD140A | MGC74795 | PDGFR2 | Rhe-PDGFRA | platelet-derived growth factor receptor, alpha polypeptide | Affinity Capture-Western | BioGRID | 8943348 |
| PDGFRB | CD140B | JTK12 | PDGF-R-beta | PDGFR | PDGFR1 | platelet-derived growth factor receptor, beta polypeptide | - | HPRD,BioGRID | 7935391 |
| PIK3C2B | C2-PI3K | DKFZp686G16234 | phosphoinositide-3-kinase, class 2, beta polypeptide | - | HPRD,BioGRID | 11533253 |
| PIK3R1 | GRB1 | p85 | p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | p85 interacts with Grb2. | BIND | 8662998 |
| PIK3R1 | GRB1 | p85 | p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | - | HPRD,BioGRID | 7642542 |7737969 |7759531 |
| PIK3R2 | P85B | p85 | p85-BETA | phosphoinositide-3-kinase, regulatory subunit 2 (beta) | - | HPRD | 8662998 |
| PLCG1 | PLC-II | PLC1 | PLC148 | PLCgamma1 | phospholipase C, gamma 1 | - | HPRD,BioGRID | 9281317 |
| PLEC1 | EBS1 | EBSO | HD1 | PCN | PLEC1b | PLTN | plectin 1, intermediate filament binding protein 500kDa | Affinity Capture-MS | BioGRID | 12577067 |
| PRKAR1A | CAR | CNC | CNC1 | DKFZp779L0468 | MGC17251 | PKR1 | PPNAD1 | PRKAR1 | TSE1 | protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) | - | HPRD,BioGRID | 9050991 |
| PRNP | ASCR | CD230 | CJD | GSS | MGC26679 | PRIP | PrP | PrP27-30 | PrP33-35C | PrPc | prion | prion protein | - | HPRD | 11571277 |
| PSMD10 | dJ889N15.2 | p28 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 | - | HPRD | 9009162 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | Tyrosine-phosphorylated FAK interacts with Grb2. | BIND | 7597091 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | - | HPRD | 7997267 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | Affinity Capture-Western Reconstituted Complex | BioGRID | 8649427 |10085298 |10806474 |12005431 |12558988 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | Grb2 interacts with FAK. This interaction was modeled on a demonstrated interaction between human Grb2 and mouse FAK. | BIND | 9148935 |
| PTK2B | CADTK | CAKB | FADK2 | FAK2 | FRNK | PKB | PTK | PYK2 | RAFTK | PTK2B protein tyrosine kinase 2 beta | - | HPRD,BioGRID | 10329689 |10354709 |
| PTPN1 | PTP1B | protein tyrosine phosphatase, non-receptor type 1 | - | HPRD | 10660596 |12388170 |
| PTPN1 | PTP1B | protein tyrosine phosphatase, non-receptor type 1 | Reconstituted Complex | BioGRID | 8940134 |10660596 |
| PTPN11 | BPTP3 | CFC | MGC14433 | NS1 | PTP-1D | PTP2C | SH-PTP2 | SH-PTP3 | SHP2 | protein tyrosine phosphatase, non-receptor type 11 | Affinity Capture-Western Far Western Reconstituted Complex | BioGRID | 8041791 |8702859 |8995399 |9362449 |9632781 |9824671 |10080542 |10212213 |10747947 |
| PTPN11 | BPTP3 | CFC | MGC14433 | NS1 | PTP-1D | PTP2C | SH-PTP2 | SH-PTP3 | SHP2 | protein tyrosine phosphatase, non-receptor type 11 | - | HPRD | 8041791 |8702859 |12531430 |
| PTPN11 | BPTP3 | CFC | MGC14433 | NS1 | PTP-1D | PTP2C | SH-PTP2 | SH-PTP3 | SHP2 | protein tyrosine phosphatase, non-receptor type 11 | Grb2 interacts with Syp via Syp SH2 domains. | BIND | 7523381 |8195176 |
| PTPN12 | PTP-PEST | PTPG1 | protein tyrosine phosphatase, non-receptor type 12 | - | HPRD,BioGRID | 9135065 |
| PTPN22 | LYP | Lyp1 | Lyp2 | PEP | PTPN8 | protein tyrosine phosphatase, non-receptor type 22 (lymphoid) | - | HPRD,BioGRID | 11882361 |
| PTPN6 | HCP | HCPH | HPTP1C | PTP-1C | SH-PTP1 | SHP-1 | SHP-1L | SHP1 | protein tyrosine phosphatase, non-receptor type 6 | - | HPRD,BioGRID | 11964172 |
| PTPRA | HEPTP | HLPR | HPTPA | HPTPalpha | LRP | PTPA | PTPRL2 | R-PTP-alpha | RPTPA | protein tyrosine phosphatase, receptor type, A | An unspecified isoform of GRB2 interacts with PTPRA. | BIND | 8670803 |
| PTPRA | HEPTP | HLPR | HPTPA | HPTPalpha | LRP | PTPA | PTPRL2 | R-PTP-alpha | RPTPA | protein tyrosine phosphatase, receptor type, A | - | HPRD,BioGRID | 7518772 |8670803 |11923305 |
| PTPRC | B220 | CD45 | CD45R | GP180 | LCA | LY5 | T200 | protein tyrosine phosphatase, receptor type, C | - | HPRD | 8570203 |
| PTPRE | DKFZp313F1310 | FLJ57799 | FLJ58245 | HPTPE | PTPE | R-PTP-EPSILON | protein tyrosine phosphatase, receptor type, E | - | HPRD,BioGRID | 10490839 |
| PXN | FLJ16691 | paxillin | Reconstituted Complex | BioGRID | 10085298 |
| RACGAP1 | HsCYK-4 | ID-GAP | MgcRacGAP | Rac GTPase activating protein 1 | Affinity Capture-MS | BioGRID | 12577067 |
| RALGPS1 | KIAA0351 | RALGEF2 | RALGPS1A | Ral GEF with PH domain and SH3 binding motif 1 | - | HPRD,BioGRID | 10747847 |
| RAPGEF1 | C3G | DKFZp781P1719 | GRF2 | Rap guanine nucleotide exchange factor (GEF) 1 | - | HPRD,BioGRID | 8621483 |
| RAPSN | CMS1D | CMS1E | MGC3597 | RNF205 | receptor-associated protein of the synapse | - | HPRD | 9856458 |
| RASA1 | CM-AVM | CMAVM | DKFZp434N071 | GAP | PKWS | RASA | RASGAP | p120GAP | p120RASGAP | RAS p21 protein activator (GTPase activating protein) 1 | - | HPRD,BioGRID | 12441060 |
| REPS2 | POB1 | RALBP1 associated Eps domain containing 2 | - | HPRD,BioGRID | 9422736 |
| RET | CDHF12 | HSCR1 | MEN2A | MEN2B | MTC1 | PTC | RET-ELE1 | RET51 | ret proto-oncogene | - | HPRD | 9393871 |
| RET | CDHF12 | HSCR1 | MEN2A | MEN2B | MTC1 | PTC | RET-ELE1 | RET51 | ret proto-oncogene | Reconstituted Complex | BioGRID | 7665556 |8183561 |
| RHOU | ARHU | CDC42L1 | DJ646B12.2 | FLJ10616 | WRCH1 | fJ646B12.2 | hG28K | ras homolog gene family, member U | Wrch-1 interacts with Grb2. This interaction was modeled on a demonstrated interaction between human Wrch-1 and Grb2 from an unspecified species. | BIND | 15556869 |
| SELL | CD62L | LAM-1 | LAM1 | LECAM1 | LNHR | LSEL | LYAM1 | Leu-8 | Lyam-1 | PLNHR | TQ1 | hLHRc | selectin L | - | HPRD,BioGRID | 8986819 |
| SGCA | 50-DAG | A2 | ADL | DAG2 | DMDA2 | LGMD2D | SCARMD1 | adhalin | sarcoglycan, alpha (50kDa dystrophin-associated glycoprotein) | - | HPRD | 7744812 |
| SH2B1 | DKFZp547G1110 | FLJ30542 | KIAA1299 | SH2-B | SH2B | SH2B adaptor protein 1 | - | HPRD,BioGRID | 9856458 |
| SH2B2 | APS | SH2B adaptor protein 2 | Affinity Capture-Western | BioGRID | 9856458 |
| SH2B3 | CELIAC13 | LNK | SH2B adaptor protein 3 | - | HPRD,BioGRID | 8649391 |
| SH3BP2 | 3BP2 | CRBM | CRPM | FLJ42079 | RES4-23 | SH3-domain binding protein 2 | - | HPRD,BioGRID | 9846481 |
| SH3KBP1 | CIN85 | GIG10 | MIG18 | SH3-domain kinase binding protein 1 | - | HPRD | 9507006|11152963 |
| SH3KBP1 | CIN85 | GIG10 | MIG18 | SH3-domain kinase binding protein 1 | SETA interacts with Grb2. This interaction was modelled on a demonstrated interaction between rat SETA and human Grb2 | BIND | 11152963 |
| SH3KBP1 | CIN85 | GIG10 | MIG18 | SH3-domain kinase binding protein 1 | Reconstituted Complex | BioGRID | 10921882 |11152963 |
| SHB | RP11-3J10.8 | bA3J10.2 | Src homology 2 domain containing adaptor protein B | - | HPRD,BioGRID | 9484780 |10488157 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | Affinity Capture-MS Affinity Capture-Western Co-fractionation Reconstituted Complex | BioGRID | 7514169 |7524086 |7535773 |8112292 |8195171 |8294403 |8491186 |8647288 |8662733 |9045692 |9083103 |9927207 |10080542 |10477741 |10570290 |11302736 |11505033 |11964172 |12441060 |12577067 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | Shc interacts with Grb2. This interaction was modelled on a demonstrated interaction between Shc from human and Grb2 from rat and an unspecified species. | BIND | 8084588 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | - | HPRD | 7524086 |7535773 |8491186 |11707405 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | An unspecified isoform of Grb2 interacts with the p52 isoform of Shc. | BIND | 9148935 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | Grb2 interacts with Shc. | BIND | 7515480 |8112292 |8939605 |10891441 |
| SHC3 | N-Shc | NSHC | RAI | SHCC | SHC (Src homology 2 domain containing) transforming protein 3 | An unspecified isoform of GRB2 interacts with SHC3 (ShcC). | BIND | 15735675 |
| SHKBP1 | PP203 | Sb1 | SH3KBP1 binding protein 1 | - | HPRD,BioGRID | 11152963 |
| SIGLEC7 | AIRM1 | CD328 | CDw328 | D-siglec | QA79 | SIGLEC-7 | p75 | p75/AIRM1 | sialic acid binding Ig-like lectin 7 | - | HPRD | 8188688 |
| SIT1 | MGC125908 | MGC125909 | MGC125910 | RP11-331F9.5 | SIT | signaling threshold regulating transmembrane adaptor 1 | - | HPRD | 11433379 |
| SKAP1 | SCAP1 | SKAP55 | src kinase associated phosphoprotein 1 | - | HPRD,BioGRID | 12171928 |
| SNTA1 | SNT1 | TACIP1 | dJ1187J4.5 | syntrophin, alpha 1 (dystrophin-associated protein A1, 59kDa, acidic component) | - | HPRD,BioGRID | 11278583 |11551227 |
| SOCS1 | CIS1 | CISH1 | JAB | SOCS-1 | SSI-1 | SSI1 | TIP3 | suppressor of cytokine signaling 1 | Socs1 interacts with Grb2. This interaction was modeled on a demonstrated interaction between mouse Socs1 and human Grb2. | BIND | 10022833 |
| SOCS1 | CIS1 | CISH1 | JAB | SOCS-1 | SSI-1 | SSI1 | TIP3 | suppressor of cytokine signaling 1 | - | HPRD,BioGRID | 10022833 |
| SOCS7 | NAP4 | suppressor of cytokine signaling 7 | - | HPRD | 9344857 |
| SOS1 | GF1 | GGF1 | GINGF | HGF | NS4 | son of sevenless homolog 1 (Drosophila) | Grb2 interacts with Sos1. This interaction was modelled on a demonstrated interaction between rat Grb2 and Sos1. | BIND | 8112292 |
| SOS1 | GF1 | GGF1 | GINGF | HGF | NS4 | son of sevenless homolog 1 (Drosophila) | - | HPRD | 8479541 |8493579 |
| SOS1 | GF1 | GGF1 | GINGF | HGF | NS4 | son of sevenless homolog 1 (Drosophila) | Affinity Capture-MS Affinity Capture-Western Far Western Reconstituted Complex Two-hybrid | BioGRID | 7510700 |7514169 |7629168 |7664271 |8112292 |8188688 |8479541 |8695800 |8810325 |9009162 |9344843 |9872323 |10207103 |10319320 |10477741 |10570290 |10716926 |11960376 |11964172 |11997436 |12577067 |
| SOS1 | GF1 | GGF1 | GINGF | HGF | NS4 | son of sevenless homolog 1 (Drosophila) | GRB2 interacts with SOS. This interaction was modeled on a demonstrated interaction between human GRB2 and mouse SOS. | BIND | 7862111 |
| SOS1 | GF1 | GGF1 | GINGF | HGF | NS4 | son of sevenless homolog 1 (Drosophila) | Sos1 interacts with Grb2. This interaction was modeled on a demonstrated interaction between Sos1 and Grb2 from Bos taurus. | BIND | 9374537 |
| SOS2 | FLJ25596 | son of sevenless homolog 2 (Drosophila) | - | HPRD,BioGRID | 7629138 |10940929 |
| SPRY1 | hSPRY1 | sprouty homolog 1, antagonist of FGF signaling (Drosophila) | - | HPRD | 12402043 |
| SPRY2 | MGC23039 | hSPRY2 | sprouty homolog 2 (Drosophila) | - | HPRD,BioGRID | 10940627 |12402043 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | - | HPRD,BioGRID | 11964172 |
| SYK | DKFZp313N1010 | FLJ25043 | FLJ37489 | spleen tyrosine kinase | - | HPRD,BioGRID | 11964172 |
| SYN1 | SYN1a | SYN1b | SYNI | synapsin I | - | HPRD | 8022809 |
| SYN1 | SYN1a | SYN1b | SYNI | synapsin I | - | HPRD,BioGRID | 8022809|10899172 |
| SYNCRIP | GRY-RBP | HNRPQ1 | NSAP1 | RP1-3J17.2 | dJ3J17.2 | hnRNP-Q | pp68 | synaptotagmin binding, cytoplasmic RNA interacting protein | Reconstituted Complex | BioGRID | 9341187 |
| SYNJ2 | INPP5H | KIAA0348 | MGC44422 | synaptojanin 2 | - | HPRD | 9388224 |
| SYP | - | synaptophysin | Affinity Capture-Western | BioGRID | 7534299 |
| TEK | CD202B | TIE-2 | TIE2 | VMCM | VMCM1 | TEK tyrosine kinase, endothelial | - | HPRD | 7478529 |10521483 |
| TFAP2A | AP-2 | AP-2alpha | AP2TF | BOFS | TFAP2 | transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) | Affinity Capture-MS | BioGRID | 12577067 |
| TFAP2B | AP-2B | AP2-B | MGC21381 | transcription factor AP-2 beta (activating enhancer binding protein 2 beta) | Affinity Capture-MS | BioGRID | 12577067 |
| TNFRSF1A | CD120a | FPF | MGC19588 | TBP1 | TNF-R | TNF-R-I | TNF-R55 | TNFAR | TNFR1 | TNFR55 | TNFR60 | p55 | p55-R | p60 | tumor necrosis factor receptor superfamily, member 1A | - | HPRD,BioGRID | 10359574 |
| TNK2 | ACK | ACK1 | FLJ44758 | FLJ45547 | p21cdc42Hs | tyrosine kinase, non-receptor, 2 | - | HPRD,BioGRID | 8647288 |
| TOM1L1 | OK/KNS-CL.3 | SRCASM | target of myb1 (chicken)-like 1 | - | HPRD,BioGRID | 11711534 |
| TP63 | AIS | B(p51A) | B(p51B) | EEC3 | KET | LMS | NBP | OFC8 | RHS | SHFM4 | TP53CP | TP53L | TP73L | p40 | p51 | p53CP | p63 | p73H | p73L | tumor protein p63 | - | HPRD | 8695800 |
| TRAT1 | HSPC062 | TCRIM | TRIM | T cell receptor associated transmembrane adaptor 1 | Reconstituted Complex | BioGRID | 10790433 |
| TUB | rd5 | tubby homolog (mouse) | - | HPRD | 10455176 |
| USP6NL | KIAA0019 | RNTRE | TRE2NL | USP6 N-terminal like | Grb2 interacts with RN-tre. | BIND | 12399475 |
| USP6NL | KIAA0019 | RNTRE | TRE2NL | USP6 N-terminal like | - | HPRD,BioGRID | 12399475 |
| USP8 | FLJ34456 | HumORF8 | KIAA0055 | MGC129718 | UBPY | ubiquitin specific peptidase 8 | - | HPRD,BioGRID | 10982817 |13129930 |
| VAV1 | VAV | vav 1 guanine nucleotide exchange factor | - | HPRD,BioGRID | 7809090 |
| VAV1 | VAV | vav 1 guanine nucleotide exchange factor | Grb2 interacts with Vav | BIND | 9013873 |
| VAV2 | - | vav 2 guanine nucleotide exchange factor | - | HPRD,BioGRID | 11606575 |
| VAV3 | FLJ40431 | vav 3 guanine nucleotide exchange factor | - | HPRD,BioGRID | 11094073 |
| WAS | IMD2 | THC | WASP | Wiskott-Aldrich syndrome (eczema-thrombocytopenia) | Affinity Capture-Western Reconstituted Complex | BioGRID | 8805332 |9307968 |
| WAS | IMD2 | THC | WASP | Wiskott-Aldrich syndrome (eczema-thrombocytopenia) | - | HPRD | 9307968 |10224664 |
| WASF1 | FLJ31482 | KIAA0269 | SCAR1 | WAVE | WAVE1 | WAS protein family, member 1 | - | HPRD | 9307968 |
| WASF2 | SCAR2 | WAVE2 | dJ393P12.2 | WAS protein family, member 2 | - | HPRD | 9307968 |
| WASL | DKFZp779G0847 | MGC48327 | N-WASP | NWASP | Wiskott-Aldrich syndrome-like | - | HPRD,BioGRID | 10087612 |10781580 |
| ZAP70 | FLJ17670 | FLJ17679 | SRK | STD | TZK | ZAP-70 | zeta-chain (TCR) associated protein kinase 70kDa | Affinity Capture-Western | BioGRID | 7629168 |
Section V. Pathway annotation
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124/506 | 996 | 1002 | m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-128 | 2260 | 2266 | 1A | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-141/200a | 2190 | 2196 | m8 | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-153 | 375 | 382 | 1A,m8 | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-182 | 183 | 189 | 1A | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-27 | 2260 | 2267 | 1A,m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-320 | 1232 | 1239 | 1A,m8 | hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA |
| miR-329 | 2165 | 2171 | 1A | hsa-miR-329brain | AACACACCUGGUUAACCUCUUU |
| miR-376c | 2208 | 2214 | 1A | hsa-miR-376c | AACAUAGAGGAAAUUCCACG |
| miR-433-3p | 2137 | 2144 | 1A,m8 | hsa-miR-433brain | AUCAUGAUGGGCUCCUCGGUGU |
| miR-448 | 376 | 382 | 1A | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-96 | 183 | 189 | 1A | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.