Gene Page: GRIA1
Summary ?
| GeneID | 2890 |
| Symbol | GRIA1 |
| Synonyms | GLUH1|GLUR1|GLURA|GluA1|HBGR1 |
| Description | glutamate ionotropic receptor AMPA type subunit 1 |
| Reference | MIM:138248|HGNC:HGNC:4571|Ensembl:ENSG00000155511|HPRD:00695|Vega:OTTHUMG00000130148 |
| Gene type | protein-coding |
| Map location | 5q31.1 |
| Pascal p-value | 0.197 |
| Fetal beta | -0.218 |
| eGene | Myers' cis & trans |
| Support | LIGAND GATED ION SIGNALING G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_TAP-PSD-95-CORE G2Cdb.humanARC G2Cdb.humanPSD G2Cdb.humanPSP Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 1 | Link to SZGene |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0032 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01718 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00459 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 5 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.4005 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4958734 | chr5 | 153789504 | GRIA1 | 2890 | 0.1 | cis | ||
| rs6712415 | chr2 | 133868796 | GRIA1 | 2890 | 0.05 | trans | ||
| rs10184338 | chr2 | 133868936 | GRIA1 | 2890 | 0.07 | trans | ||
| rs10172235 | chr2 | 133869155 | GRIA1 | 2890 | 0.17 | trans | ||
| rs17029291 | chr3 | 32402138 | GRIA1 | 2890 | 8.566E-5 | trans | ||
| rs3988303 | chr8 | 19776538 | GRIA1 | 2890 | 0.15 | trans | ||
| rs11793484 | chr9 | 317239 | GRIA1 | 2890 | 0.03 | trans | ||
| rs10970457 | chr9 | 326183 | GRIA1 | 2890 | 0.03 | trans |
Section II. Transcriptome annotation
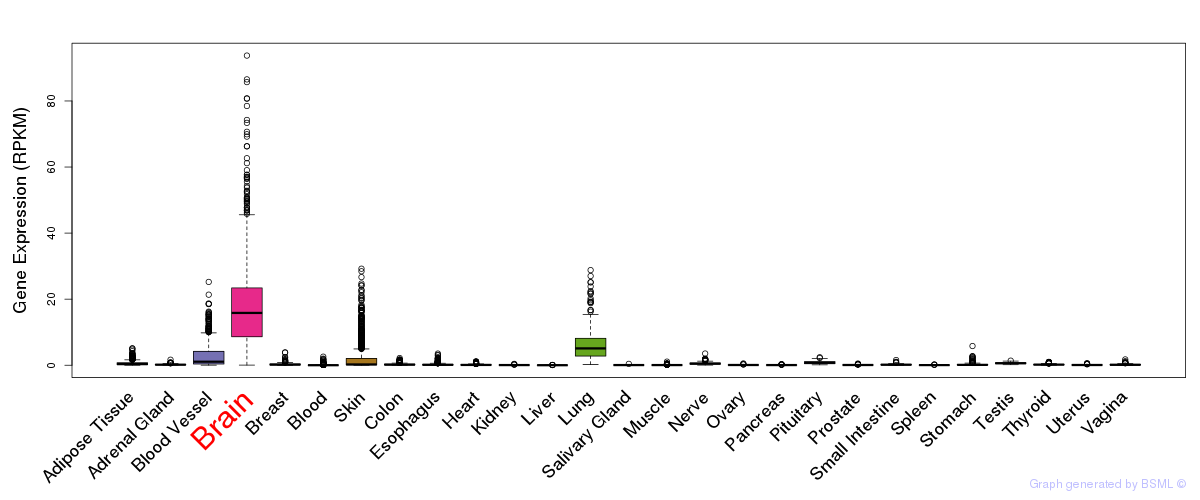
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PORCN | 0.90 | 0.87 |
| C3orf39 | 0.89 | 0.88 |
| SCYL1 | 0.89 | 0.87 |
| FAM127A | 0.88 | 0.86 |
| ATP6AP1 | 0.88 | 0.87 |
| RNF123 | 0.87 | 0.83 |
| ACP2 | 0.86 | 0.85 |
| SLC45A1 | 0.86 | 0.87 |
| CDIPT | 0.86 | 0.86 |
| C17orf28 | 0.85 | 0.82 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.58 | -0.44 |
| AF347015.18 | -0.56 | -0.48 |
| NSBP1 | -0.56 | -0.55 |
| AL139819.3 | -0.53 | -0.51 |
| MT-ATP8 | -0.52 | -0.42 |
| AF347015.8 | -0.52 | -0.39 |
| GNG11 | -0.52 | -0.45 |
| MT-CO2 | -0.50 | -0.37 |
| NOSTRIN | -0.50 | -0.39 |
| AF347015.31 | -0.50 | -0.37 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005234 | extracellular-glutamate-gated ion channel activity | IEA | glutamate (GO term level: 11) | - |
| GO:0015277 | kainate selective glutamate receptor activity | TAS | glutamate (GO term level: 8) | 1311100 |
| GO:0004872 | receptor activity | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0005216 | ion channel activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007268 | synaptic transmission | TAS | neuron, Synap, Neurotransmitter (GO term level: 6) | 1311100 |
| GO:0007165 | signal transduction | TAS | 1311100 | |
| GO:0007616 | long-term memory | IEA | - | |
| GO:0006811 | ion transport | IEA | - | |
| GO:0031623 | receptor internalization | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0014069 | postsynaptic density | IEA | Synap (GO term level: 10) | - |
| GO:0045211 | postsynaptic membrane | IEA | Synap, Neurotransmitter (GO term level: 5) | - |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0005789 | endoplasmic reticulum membrane | IEA | - | |
| GO:0005624 | membrane fraction | IEA | - | |
| GO:0005783 | endoplasmic reticulum | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | TAS | 1311100 | |
| GO:0030054 | cell junction | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CACNG2 | MGC138502 | MGC138504 | calcium channel, voltage-dependent, gamma subunit 2 | - | HPRD,BioGRID | 11140673 |
| CANX | CNX | FLJ26570 | IP90 | P90 | calnexin | - | HPRD | 10461883 |
| DLG1 | DKFZp761P0818 | DKFZp781B0426 | DLGH1 | SAP97 | dJ1061C18.1.1 | hdlg | discs, large homolog 1 (Drosophila) | - | HPRD,BioGRID | 9677374 |11567040 |
| EPB41L1 | 4.1N | DKFZp686H17242 | KIAA0338 | MGC11072 | erythrocyte membrane protein band 4.1-like 1 | - | HPRD,BioGRID | 11050113 |
| EPB41L2 | 4.1-G | DKFZp781D1972 | DKFZp781H1755 | erythrocyte membrane protein band 4.1-like 2 | - | HPRD,BioGRID | 11050113 |
| GRIA2 | GLUR2 | GLURB | GluR-K2 | HBGR2 | glutamate receptor, ionotropic, AMPA 2 | Affinity Capture-Western | BioGRID | 10358037 |
| GRIA4 | GLUR4 | GLUR4C | GLURD | glutamate receptor, ionotrophic, AMPA 4 | - | HPRD,BioGRID | 9466455 |
| GRID2 | MGC117022 | MGC117023 | MGC117024 | glutamate receptor, ionotropic, delta 2 | Affinity Capture-Western | BioGRID | 12573530 |
| GRID2 | MGC117022 | MGC117023 | MGC117024 | glutamate receptor, ionotropic, delta 2 | GluR1 interacts with GluR-delta-2 | BIND | 12573530 |
| GRIK2 | EAA4 | GLR6 | GLUK6 | GLUR6 | MGC74427 | MRT6 | glutamate receptor, ionotropic, kainate 2 | Affinity Capture-Western | BioGRID | 8288598 |
| GRIK2 | EAA4 | GLR6 | GLUK6 | GLUR6 | MGC74427 | MRT6 | glutamate receptor, ionotropic, kainate 2 | - | HPRD | 10358037 |
| GRIP1 | GRIP | glutamate receptor interacting protein 1 | Reconstituted Complex | BioGRID | 11891216 |
| HOMER1 | HOMER | HOMER1A | HOMER1B | HOMER1C | SYN47 | Ves-1 | homer homolog 1 (Drosophila) | - | HPRD,BioGRID | 9069287 |9808458 |11418862 |
| HSPA5 | BIP | FLJ26106 | GRP78 | MIF2 | heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) | - | HPRD | 10461883 |
| PICK1 | MGC15204 | PICK | PRKCABP | protein interacting with PRKCA 1 | - | HPRD,BioGRID | 11891216 |
| SDCBP | MDA-9 | ST1 | SYCL | TACIP18 | syndecan binding protein (syntenin) | - | HPRD,BioGRID | 11891216 |
| TANC1 | KIAA1728 | ROLSB | TANC | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 | - | HPRD | 15673434 |
| TRPC1 | HTRP-1 | MGC133334 | MGC133335 | TRP1 | transient receptor potential cation channel, subfamily C, member 1 | - | HPRD | 14614461 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| KEGG LONG TERM POTENTIATION | 70 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG LONG TERM DEPRESSION | 70 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG AMYOTROPHIC LATERAL SCLEROSIS ALS | 53 | 43 | All SZGR 2.0 genes in this pathway |
| PID EPHB FWD PATHWAY | 40 | 38 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL | 137 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAFFICKING OF AMPA RECEPTORS | 28 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | 16 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF NMDA RECEPTOR UPON GLUTAMATE BINDING AND POSTSYNAPTIC EVENTS | 37 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | 15 | 14 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LPS UP | 431 | 237 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION UP | 195 | 138 | All SZGR 2.0 genes in this pathway |
| LEE TARGETS OF PTCH1 AND SUFU DN | 83 | 69 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| MODY HIPPOCAMPUS POSTNATAL | 63 | 50 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN DN | 153 | 120 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| JIANG AGING HYPOTHALAMUS DN | 40 | 31 | All SZGR 2.0 genes in this pathway |
| MCCLUNG COCAINE REWARD 5D | 79 | 62 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 8HR UP | 105 | 73 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR UP | 180 | 125 | All SZGR 2.0 genes in this pathway |
| XU GH1 AUTOCRINE TARGETS UP | 268 | 157 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE DN | 264 | 159 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 1HR UP | 61 | 44 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS DN | 229 | 135 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL DN | 308 | 187 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |