Gene Page: GIT1
Summary ?
| GeneID | 28964 |
| Symbol | GIT1 |
| Synonyms | - |
| Description | GIT ArfGAP 1 |
| Reference | MIM:608434|HGNC:HGNC:4272|Ensembl:ENSG00000108262|HPRD:06577|Vega:OTTHUMG00000132730 |
| Gene type | protein-coding |
| Map location | 17p11.2 |
| Pascal p-value | 0.776 |
| Sherlock p-value | 0.303 |
| Fetal beta | -0.688 |
| DMG | 1 (# studies) |
| Support | ENDOCYTOSIS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| GIT1 | chr17 | 27902699 | C | T | NM_001085454 NM_014030 | p.601S>N p.592S>N | missense missense | Schizophrenia | DNM:Fromer_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg22120948 | 17 | 27915972 | GIT1 | 3.82E-9 | -0.01 | 2.4E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
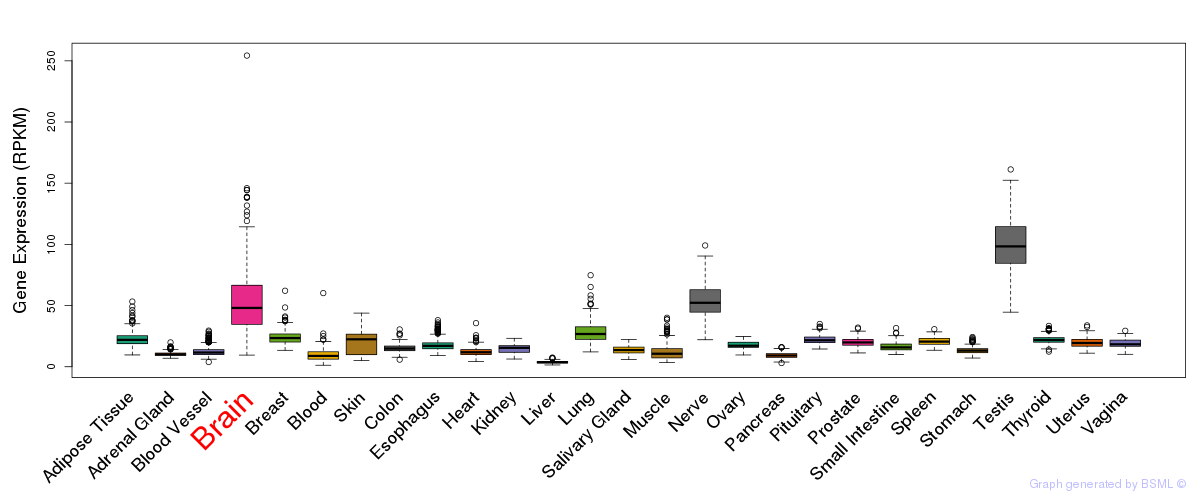
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ADRBK1 | BARK1 | BETA-ARK1 | FLJ16718 | GRK2 | adrenergic, beta, receptor kinase 1 | - | HPRD,BioGRID | 9826657 |
| ADRBK2 | BARK2 | GRK3 | adrenergic, beta, receptor kinase 2 | - | HPRD,BioGRID | 9826657 |
| ARHGEF6 | COOL2 | Cool-2 | KIAA0006 | MRX46 | PIXA | alpha-PIX | alphaPIX | Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 | - | HPRD | 10896954 |
| ARHGEF6 | COOL2 | Cool-2 | KIAA0006 | MRX46 | PIXA | alpha-PIX | alphaPIX | Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 | Affinity Capture-Western | BioGRID | 10428811 |
| ARHGEF7 | BETA-PIX | COOL1 | DKFZp686C12170 | DKFZp761K1021 | KIAA0142 | KIAA0412 | Nbla10314 | P50 | P50BP | P85 | P85COOL1 | P85SPR | PAK3 | PIXB | Rho guanine nucleotide exchange factor (GEF) 7 | Affinity Capture-Western Two-hybrid | BioGRID | 10428811 |12473661 |
| ARHGEF7 | BETA-PIX | COOL1 | DKFZp686C12170 | DKFZp761K1021 | KIAA0142 | KIAA0412 | Nbla10314 | P50 | P50BP | P85 | P85COOL1 | P85SPR | PAK3 | PIXB | Rho guanine nucleotide exchange factor (GEF) 7 | - | HPRD | 10896954 |
| ARS2 | ASR2 | MGC126427 | arsenate resistance protein 2 | Two-hybrid | BioGRID | 16169070 |
| C8orf33 | FLJ20989 | chromosome 8 open reading frame 33 | Two-hybrid | BioGRID | 16169070 |
| CCDC113 | DKFZp434N1418 | FLJ23555 | HSPC065 | coiled-coil domain containing 113 | Two-hybrid | BioGRID | 16169070 |
| DDX24 | - | DEAD (Asp-Glu-Ala-Asp) box polypeptide 24 | Two-hybrid | BioGRID | 16169070 |
| EIF6 | 2 | CAB | EIF3A | ITGB4BP | b | b(2)gcn | gcn | p27BBP | eukaryotic translation initiation factor 6 | Two-hybrid | BioGRID | 16169070 |
| ERC2 | CAST | CAST1 | ELKSL | KIAA0378 | MGC133063 | MGC133064 | SPBC110 | Spc110 | ELKS/RAB6-interacting/CAST family member 2 | - | HPRD,BioGRID | 12923177 |
| GIT1 | - | G protein-coupled receptor kinase interacting ArfGAP 1 | - | HPRD,BioGRID | 12473661 |
| GIT2 | CAT-2 | DKFZp686G01261 | KIAA0148 | MGC760 | G protein-coupled receptor kinase interacting ArfGAP 2 | - | HPRD,BioGRID | 12473661 |
| GRIP2 | - | glutamate receptor interacting protein 2 | Affinity Capture-Western | BioGRID | 12629171 |
| GRK5 | GPRK5 | G protein-coupled receptor kinase 5 | - | HPRD | 9826657 |
| GRK6 | FLJ32135 | GPRK6 | G protein-coupled receptor kinase 6 | - | HPRD | 9826657 |
| HBXIP | MGC71071 | XIP | hepatitis B virus x interacting protein | Two-hybrid | BioGRID | 16169070 |
| HMOX2 | HO-2 | heme oxygenase (decycling) 2 | Two-hybrid | BioGRID | 16169070 |
| KIF1A | ATSV | C2orf20 | DKFZp686I2094 | FLJ30229 | HUNC-104 | MGC133285 | MGC133286 | UNC104 | kinesin family member 1A | - | HPRD | 12522103 |
| MAN2A2 | MANA2X | mannosidase, alpha, class 2A, member 2 | Two-hybrid | BioGRID | 16169070 |
| PAK2 | PAK65 | PAKgamma | p21 protein (Cdc42/Rac)-activated kinase 2 | Affinity Capture-MS | BioGRID | 17353931 |
| PAK3 | CDKN1A | MRX30 | MRX47 | OPHN3 | PAK3beta | bPAK | hPAK3 | p21 protein (Cdc42/Rac)-activated kinase 3 | - | HPRD | 10896954 |
| PCLO | ACZ | DKFZp779G1236 | piccolo (presynaptic cytomatrix protein) | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 12473661 |
| PFDN1 | PDF | PFD1 | prefoldin subunit 1 | Two-hybrid | BioGRID | 16169070 |
| PLCG1 | PLC-II | PLC1 | PLC148 | PLCgamma1 | phospholipase C, gamma 1 | - | HPRD,BioGRID | 14523024 |
| PMF1 | - | polyamine-modulated factor 1 | Two-hybrid | BioGRID | 16169070 |
| PPFIA1 | FLJ41337 | FLJ42630 | FLJ43474 | LIP.1 | LIP1 | LIPRIN | MGC26800 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1 | Liprin-alpha1 interacts with GIT1. This interaction was modeled on a demonstrated interaction between human liprin-alpha1 and rat GIT1. | BIND | 12923177 |
| PPFIA1 | FLJ41337 | FLJ42630 | FLJ43474 | LIP.1 | LIP1 | LIPRIN | MGC26800 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1 | - | HPRD,BioGRID | 12629171 |
| PPFIA2 | FLJ41378 | MGC132572 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 | Reconstituted Complex Two-hybrid | BioGRID | 12923177 |
| PPFIA2 | FLJ41378 | MGC132572 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 | - | HPRD | 12629171 |
| PPFIA2 | FLJ41378 | MGC132572 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 | Liprin-alpha2 interacts with GIT1. This interaction was modeled on a demonstrated interaction between human liprin-alpha2 and rat GIT1. | BIND | 12923177 |
| PPFIA3 | KIAA0654 | LPNA3 | MGC126567 | MGC126569 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 | Reconstituted Complex Two-hybrid | BioGRID | 12923177 |
| PPFIA3 | KIAA0654 | LPNA3 | MGC126567 | MGC126569 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 | Liprin-alpha3 interacts with GIT1. This interaction was modeled on a demonstrated interaction between human liprin-alpha3 and rat GIT1. | BIND | 12923177 |
| PPFIA4 | - | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 | - | HPRD,BioGRID | 12629171 |12923177 |
| PPFIA4 | - | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 | Liprin-alpha4 interacts with GIT1. This interaction was modeled on a demonstrated interaction between human Liprin-alpha4 and rat GIT1. | BIND | 12923177 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | - | HPRD,BioGRID | 10938112 |
| PTK2B | CADTK | CAKB | FADK2 | FAK2 | FRNK | PKB | PTK | PYK2 | RAFTK | PTK2B protein tyrosine kinase 2 beta | Affinity Capture-Western | BioGRID | 12629171 |
| PTPRF | FLJ43335 | FLJ45062 | FLJ45567 | LAR | protein tyrosine phosphatase, receptor type, F | Affinity Capture-Western | BioGRID | 12629171 |
| PTPRZ1 | HPTPZ | HPTPzeta | PTP-ZETA | PTP18 | PTPRZ | PTPZ | RPTPB | RPTPbeta | phosphacan | protein tyrosine phosphatase, receptor-type, Z polypeptide 1 | Affinity Capture-Western | BioGRID | 11381105 |
| PXN | FLJ16691 | paxillin | GIT1 interacts with Paxillin (PXN). | BIND | 15793570 |
| PXN | FLJ16691 | paxillin | Affinity Capture-Western | BioGRID | 12629171 |
| PXN | FLJ16691 | paxillin | - | HPRD | 10896954 |12153727 |14523024 |
| RGS2 | G0S8 | regulator of G-protein signaling 2, 24kDa | Two-hybrid | BioGRID | 16169070 |
| SDCCAG3 | NY-CO-3 | serologically defined colon cancer antigen 3 | Two-hybrid | BioGRID | 16169070 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | - | HPRD | 14523024 |
| TERF1 | FLJ41416 | PIN2 | TRBF1 | TRF | TRF1 | hTRF1-AS | t-TRF1 | telomeric repeat binding factor (NIMA-interacting) 1 | Two-hybrid | BioGRID | 16169070 |
| TGFB1I1 | ARA55 | HIC-5 | HIC5 | TSC-5 | transforming growth factor beta 1 induced transcript 1 | - | HPRD | 12153727 |
| TRIB3 | C20orf97 | NIPK | SINK | SKIP3 | TRB3 | tribbles homolog 3 (Drosophila) | Two-hybrid | BioGRID | 16169070 |
| TXNDC9 | APACD | thioredoxin domain containing 9 | Two-hybrid | BioGRID | 16169070 |
| WDR33 | FLJ11294 | WDC146 | WD repeat domain 33 | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ENDOCYTOSIS | 183 | 132 | All SZGR 2.0 genes in this pathway |
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| KEGG EPITHELIAL CELL SIGNALING IN HELICOBACTER PYLORI INFECTION | 68 | 44 | All SZGR 2.0 genes in this pathway |
| PID CDC42 REG PATHWAY | 30 | 22 | All SZGR 2.0 genes in this pathway |
| PID ARF6 PATHWAY | 35 | 27 | All SZGR 2.0 genes in this pathway |
| PID ECADHERIN STABILIZATION PATHWAY | 42 | 34 | All SZGR 2.0 genes in this pathway |
| PID AURORA A PATHWAY | 31 | 20 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN A4B1 PATHWAY | 33 | 24 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS TOP50 UP | 38 | 27 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE DN | 384 | 220 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 17Q11 Q21 AMPLICON | 133 | 78 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| MOREAUX B LYMPHOCYTE MATURATION BY TACI UP | 92 | 58 | All SZGR 2.0 genes in this pathway |
| MARIADASON REGULATED BY HISTONE ACETYLATION DN | 54 | 30 | All SZGR 2.0 genes in this pathway |
| HUNSBERGER EXERCISE REGULATED GENES | 31 | 26 | All SZGR 2.0 genes in this pathway |