Gene Page: DBNL
Summary ?
| GeneID | 28988 |
| Symbol | DBNL |
| Synonyms | ABP1|HIP-55|HIP55|SH3P7 |
| Description | drebrin like |
| Reference | MIM:610106|HGNC:HGNC:2696|Ensembl:ENSG00000136279|HPRD:06588|Vega:OTTHUMG00000155350 |
| Gene type | protein-coding |
| Map location | 7p13 |
| Pascal p-value | 0.697 |
| Sherlock p-value | 0.204 |
| Fetal beta | -0.689 |
| DMG | 2 (# studies) |
| eGene | Myers' cis & trans Meta |
| Support | STRUCTURAL PLASTICITY CompositeSet Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg09138430 | 7 | 44084267 | DBNL | 2.557E-4 | -0.258 | 0.037 | DMG:Wockner_2014 |
| cg26086607 | 7 | 44084030 | DBNL | 5.83E-8 | -0.008 | 1.49E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs758989 | chr7 | 44203005 | DBNL | 28988 | 0.03 | cis | ||
| rs1303722 | chr7 | 44219073 | DBNL | 28988 | 0 | cis |
Section II. Transcriptome annotation
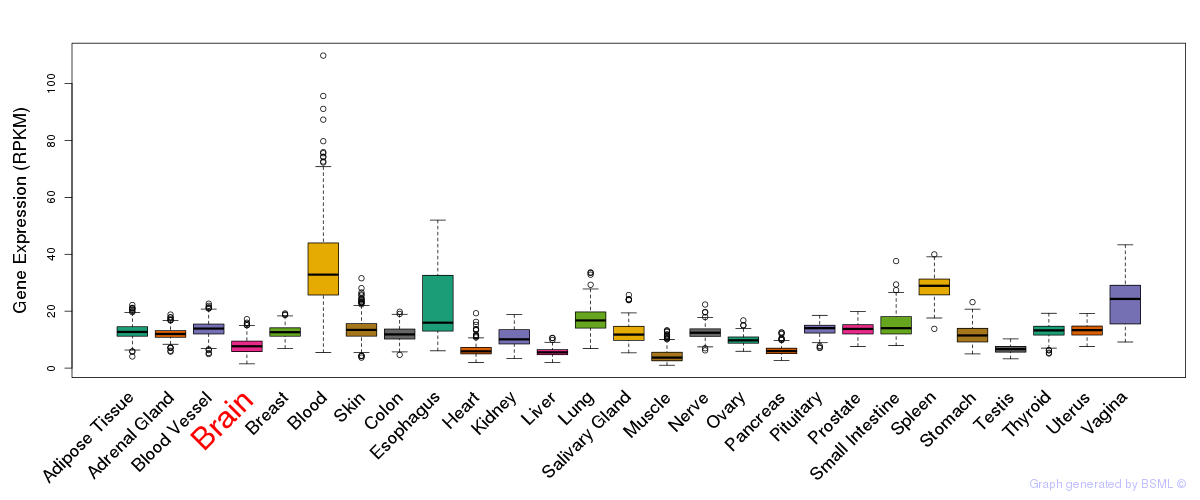
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CASP3 | CPP32 | CPP32B | SCA-1 | caspase 3, apoptosis-related cysteine peptidase | Biochemical Activity | BioGRID | 11689006 |
| CDK4 | CMM3 | MGC14458 | PSK-J3 | cyclin-dependent kinase 4 | Affinity Capture-MS | BioGRID | 17353931 |
| CYP4F8 | CPF8 | CYPIVF8 | cytochrome P450, family 4, subfamily F, polypeptide 8 | Affinity Capture-MS | BioGRID | 17353931 |
| DNPEP | ASPEP | DAP | aspartyl aminopeptidase | Affinity Capture-MS | BioGRID | 17353931 |
| EIF2AK2 | EIF2AK1 | MGC126524 | PKR | PRKR | eukaryotic translation initiation factor 2-alpha kinase 2 | Affinity Capture-MS | BioGRID | 17353931 |
| GPS2 | AMF-1 | MGC104294 | MGC119287 | MGC119288 | MGC119289 | G protein pathway suppressor 2 | Affinity Capture-MS | BioGRID | 17353931 |
| HERC2 | D15F37S1 | DKFZp547P028 | KIAA0393 | SHEP1 | jdf2 | p528 | hect domain and RLD 2 | Affinity Capture-MS | BioGRID | 17353931 |
| MAP4K1 | HPK1 | mitogen-activated protein kinase kinase kinase kinase 1 | - | HPRD,BioGRID | 10567356 |
| RB1CC1 | CC1 | DRAGOU14 | FIP200 | RB1-inducible coiled-coil 1 | Affinity Capture-MS | BioGRID | 17353931 |
| SH2D4A | FLJ20967 | SH2A | SH2 domain containing 4A | Two-hybrid | BioGRID | 16189514 |
| ZAP70 | FLJ17670 | FLJ17679 | SRK | STD | TZK | ZAP-70 | zeta-chain (TCR) associated protein kinase 70kDa | - | HPRD,BioGRID | 14557276 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID TCR PATHWAY | 66 | 51 | All SZGR 2.0 genes in this pathway |
| PID TCR JNK PATHWAY | 14 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | 40 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | 13 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOSIS | 148 | 94 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOTIC EXECUTION PHASE | 54 | 37 | All SZGR 2.0 genes in this pathway |
| MORI SMALL PRE BII LYMPHOCYTE UP | 86 | 57 | All SZGR 2.0 genes in this pathway |
| TSENG IRS1 TARGETS UP | 113 | 71 | All SZGR 2.0 genes in this pathway |
| TSENG ADIPOGENIC POTENTIAL UP | 30 | 19 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS DN | 314 | 188 | All SZGR 2.0 genes in this pathway |
| MCCABE BOUND BY HOXC6 | 469 | 239 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 13 | 172 | 107 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |