Gene Page: GRIN2B
Summary ?
| GeneID | 2904 |
| Symbol | GRIN2B |
| Synonyms | EIEE27|GluN2B|MRD6|NMDAR2B|NR2B|hNR3 |
| Description | glutamate ionotropic receptor NMDA type subunit 2B |
| Reference | MIM:138252|HGNC:HGNC:4586|Ensembl:ENSG00000273079|HPRD:00697|Vega:OTTHUMG00000137373 |
| Gene type | protein-coding |
| Map location | 12p12 |
| Pascal p-value | 0.003 |
| Fetal beta | 0.236 |
| eGene | Meta |
| Support | LIGAND GATED ION SIGNALING G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_PocklingtonH1 G2Cdb.human_TAP-PSD-95-CORE G2Cdb.humanNRC G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| ADT:Sun_2012 | Systematic Investigation of Antipsychotic Drugs and Their Targets | A total of 382 drug-target associations involving 43 antipsychotic drugs and 49 target genes. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Awadalla_2010 | Whole Exome Sequencing analysis | A survey of ~430 Mb of DNA from 401 synapse-expressed genes across all cases and 25 Mb of DNA in controls found 28 candidate DNMs. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 2 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 12 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 31.1091 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| GRIN2B | chr12 | 13720084 | T | G | GRIN2B:NM_000834:exon12:c.T2473C:p.L825L | L825V | synonymous SNV | Paranoid Schizophrenia | DNM:Awadalla_2010 |
Section II. Transcriptome annotation
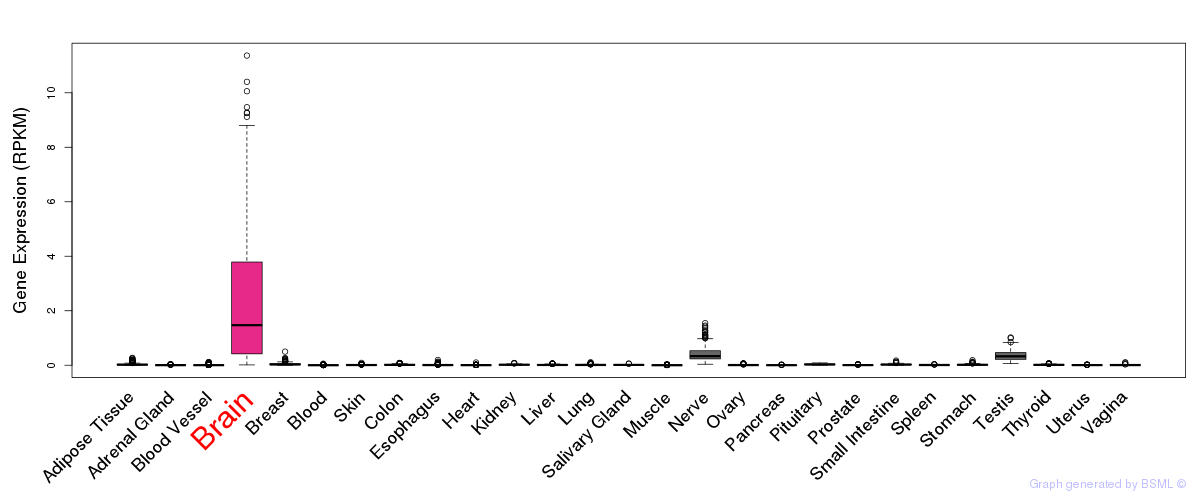
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004972 | N-methyl-D-aspartate selective glutamate receptor activity | TAS | glutamate (GO term level: 8) | 8768735 |
| GO:0005234 | extracellular-glutamate-gated ion channel activity | IEA | glutamate (GO term level: 11) | - |
| GO:0035254 | glutamate receptor binding | IEA | glutamate (GO term level: 5) | - |
| GO:0000287 | magnesium ion binding | IEA | - | |
| GO:0004872 | receptor activity | IEA | - | |
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 17047094 |17997397 | |
| GO:0005262 | calcium channel activity | IEA | - | |
| GO:0016594 | glycine binding | IDA | 17047094 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0060079 | regulation of excitatory postsynaptic membrane potential | IEA | neuron, Synap (GO term level: 10) | - |
| GO:0048167 | regulation of synaptic plasticity | IEA | Synap (GO term level: 8) | - |
| GO:0007215 | glutamate signaling pathway | TAS | glutamate (GO term level: 7) | 8768735 |
| GO:0001701 | in utero embryonic development | IEA | - | |
| GO:0001662 | behavioral fear response | IEA | - | |
| GO:0001964 | startle response | IEA | - | |
| GO:0001967 | suckling behavior | IEA | - | |
| GO:0009790 | embryonic development | IEA | - | |
| GO:0007612 | learning | IEA | - | |
| GO:0007613 | memory | IEA | - | |
| GO:0007423 | sensory organ development | IEA | - | |
| GO:0006816 | calcium ion transport | IEA | - | |
| GO:0006812 | cation transport | IEA | - | |
| GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain | IEA | - | |
| GO:0048266 | behavioral response to pain | IEA | - | |
| GO:0045471 | response to ethanol | IDA | 18445116 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0042734 | presynaptic membrane | IEA | neuron, axon, Synap (GO term level: 5) | - |
| GO:0008021 | synaptic vesicle | IEA | Synap, Neurotransmitter (GO term level: 12) | - |
| GO:0014069 | postsynaptic density | IEA | Synap (GO term level: 10) | - |
| GO:0019717 | synaptosome | IEA | Synap, Brain (GO term level: 7) | - |
| GO:0017146 | N-methyl-D-aspartate selective glutamate receptor complex | IDA | glutamate, Synap (GO term level: 10) | 10480938 |17047094 |
| GO:0045211 | postsynaptic membrane | IEA | Synap, Neurotransmitter (GO term level: 5) | - |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 8768735 | |
| GO:0030054 | cell junction | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTN2 | CMD1AA | actinin, alpha 2 | - | HPRD,BioGRID | 9009191 |
| CAMK2A | CAMKA | KIAA0968 | calcium/calmodulin-dependent protein kinase II alpha | - | HPRD,BioGRID | 11459059 |
| CAMK2N1 | MGC22256 | PRO1489 | RP11-401M16.1 | calcium/calmodulin-dependent protein kinase II inhibitor 1 | - | HPRD | 12650977 |
| DLG1 | DKFZp761P0818 | DKFZp781B0426 | DLGH1 | SAP97 | dJ1061C18.1.1 | hdlg | discs, large homolog 1 (Drosophila) | Affinity Capture-Western | BioGRID | 11997254 |
| DLG2 | DKFZp781D1854 | DKFZp781E0954 | FLJ37266 | MGC131811 | PSD-93 | discs, large homolog 2, chapsyn-110 (Drosophila) | Affinity Capture-Western Two-hybrid | BioGRID | 9278515 |11997254 |
| DLG2 | DKFZp781D1854 | DKFZp781E0954 | FLJ37266 | MGC131811 | PSD-93 | discs, large homolog 2, chapsyn-110 (Drosophila) | - | HPRD | 9806853 |11997254 |
| DLG3 | KIAA1232 | MRX | MRX90 | NE-Dlg | NEDLG | SAP102 | discs, large homolog 3 (neuroendocrine-dlg, Drosophila) | - | HPRD,BioGRID | 11937501 |
| DLG4 | FLJ97752 | FLJ98574 | PSD95 | SAP90 | discs, large homolog 4 (Drosophila) | Affinity Capture-Western in vitro in vivo Reconstituted Complex Two-hybrid | BioGRID | 7569905 |9278515 |9581762 |11937501 |11997254 |12738960 |
| DLG4 | FLJ97752 | FLJ98574 | PSD95 | SAP90 | discs, large homolog 4 (Drosophila) | - | HPRD | 7569905 |9581762|11937501 |
| ERBB2IP | ERBIN | LAP2 | erbb2 interacting protein | Reconstituted Complex | BioGRID | 11278603 |
| EXOC3 | SEC6 | SEC6L1 | Sec6p | exocyst complex component 3 | Affinity Capture-Western | BioGRID | 12738960 |
| EXOC4 | MGC27170 | REC8 | SEC8 | SEC8L1 | Sec8p | exocyst complex component 4 | - | HPRD,BioGRID | 12738960 |
| EXOC7 | 2-5-3p | DKFZp686J04253 | EX070 | EXO70 | EXOC1 | Exo70p | FLJ40965 | FLJ46415 | YJL085W | exocyst complex component 7 | Reconstituted Complex | BioGRID | 12738960 |
| FYN | MGC45350 | SLK | SYN | FYN oncogene related to SRC, FGR, YES | - | HPRD,BioGRID | 10458595 |
| GNB2L1 | Gnb2-rs1 | H12.3 | HLC-7 | PIG21 | RACK1 | guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 | - | HPRD,BioGRID | 12524444 |
| GRIN1 | NMDA1 | NMDAR1 | NR1 | glutamate receptor, ionotropic, N-methyl D-aspartate 1 | Affinity Capture-Western | BioGRID | 12391275 |
| GRIN1 | NMDA1 | NMDAR1 | NR1 | glutamate receptor, ionotropic, N-methyl D-aspartate 1 | - | HPRD | 8139656 |12191494 |15030493 |
| IL16 | FLJ16806 | FLJ42735 | FLJ44234 | HsT19289 | IL-16 | LCF | prIL-16 | interleukin 16 (lymphocyte chemoattractant factor) | - | HPRD,BioGRID | 10479680 |
| INADL | Cipp | FLJ26982 | InaD-like | PATJ | InaD-like (Drosophila) | - | HPRD | 9647694 |
| LIN7A | LIN-7A | LIN7 | MALS-1 | MGC148143 | TIP-33 | VELI1 | lin-7 homolog A (C. elegans) | in vitro | BioGRID | 10341223 |
| LIN7B | LIN-7B | MALS-2 | MALS2 | VELI2 | lin-7 homolog B (C. elegans) | in vitro in vivo | BioGRID | 10341223 |
| MAGI3 | MAGI-3 | MGC163281 | RP4-730K3.1 | dJ730K3.2 | membrane associated guanylate kinase, WW and PDZ domain containing 3 | - | HPRD | 10748157 |
| MAGI3 | MAGI-3 | MGC163281 | RP4-730K3.1 | dJ730K3.2 | membrane associated guanylate kinase, WW and PDZ domain containing 3 | MAGI3 interacts with NMDA receptor 2B. | BIND | 10748157 |
| PARK2 | AR-JP | LPRS2 | PDJ | PRKN | Parkinson disease (autosomal recessive, juvenile) 2, parkin | - | HPRD,BioGRID | 11679592 |
| RICS | GC-GAP | GRIT | KIAA0712 | MGC1892 | p200RhoGAP | p250GAP | Rho GTPase-activating protein | - | HPRD,BioGRID | 12531901 |12857875 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | - | HPRD | 10884433 |
| SYNGAP1 | DKFZp761G1421 | KIAA1938 | RASA1 | RASA5 | SYNGAP | synaptic Ras GTPase activating protein 1 homolog (rat) | - | HPRD | 9620694 |
| TANC1 | KIAA1728 | ROLSB | TANC | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 | - | HPRD | 15673434 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| KEGG LONG TERM POTENTIATION | 70 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG ALZHEIMERS DISEASE | 169 | 110 | All SZGR 2.0 genes in this pathway |
| KEGG AMYOTROPHIC LATERAL SCLEROSIS ALS | 53 | 43 | All SZGR 2.0 genes in this pathway |
| KEGG HUNTINGTONS DISEASE | 185 | 109 | All SZGR 2.0 genes in this pathway |
| KEGG SYSTEMIC LUPUS ERYTHEMATOSUS | 140 | 100 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NOS1 PATHWAY | 24 | 21 | All SZGR 2.0 genes in this pathway |
| PID ERBB4 PATHWAY | 38 | 32 | All SZGR 2.0 genes in this pathway |
| PID REELIN PATHWAY | 29 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL | 137 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | 17 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF NMDA RECEPTOR UPON GLUTAMATE BINDING AND POSTSYNAPTIC EVENTS | 37 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | 27 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | 33 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | 15 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | 15 | 14 | All SZGR 2.0 genes in this pathway |
| ROVERSI GLIOMA LOH REGIONS | 44 | 30 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| STARK HYPPOCAMPUS 22Q11 DELETION UP | 53 | 40 | All SZGR 2.0 genes in this pathway |
| CHESLER BRAIN QTL CIS | 75 | 51 | All SZGR 2.0 genes in this pathway |
| LOPES METHYLATED IN COLON CANCER DN | 28 | 26 | All SZGR 2.0 genes in this pathway |
| LEIN NEURON MARKERS | 69 | 45 | All SZGR 2.0 genes in this pathway |
| STEIN ESR1 TARGETS | 85 | 55 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K27ME3 | 341 | 243 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-204/211 | 1064 | 1071 | 1A,m8 | hsa-miR-204brain | UUCCCUUUGUCAUCCUAUGCCU |
| hsa-miR-211 | UUCCCUUUGUCAUCCUUCGCCU | ||||
| miR-330 | 1247 | 1254 | 1A,m8 | hsa-miR-330brain | GCAAAGCACACGGCCUGCAGAGA |
| miR-410 | 845 | 851 | 1A | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.