Gene Page: PYCARD
Summary ?
| GeneID | 29108 |
| Symbol | PYCARD |
| Synonyms | ASC|CARD5|TMS|TMS-1|TMS1 |
| Description | PYD and CARD domain containing |
| Reference | MIM:606838|HGNC:HGNC:16608|Ensembl:ENSG00000103490|HPRD:06020|Vega:OTTHUMG00000176753 |
| Gene type | protein-coding |
| Map location | 16p11.2 |
| Pascal p-value | 0.27 |
| Sherlock p-value | 0.274 |
| TADA p-value | 0.009 |
| Fetal beta | -1.215 |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01775 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PYCARD | chr16 | 31213117 | A | C | NM_013258 NM_145182 | p.126V>G p.107V>G | missense missense | Schizophrenia | DNM:Fromer_2014 |
Section II. Transcriptome annotation
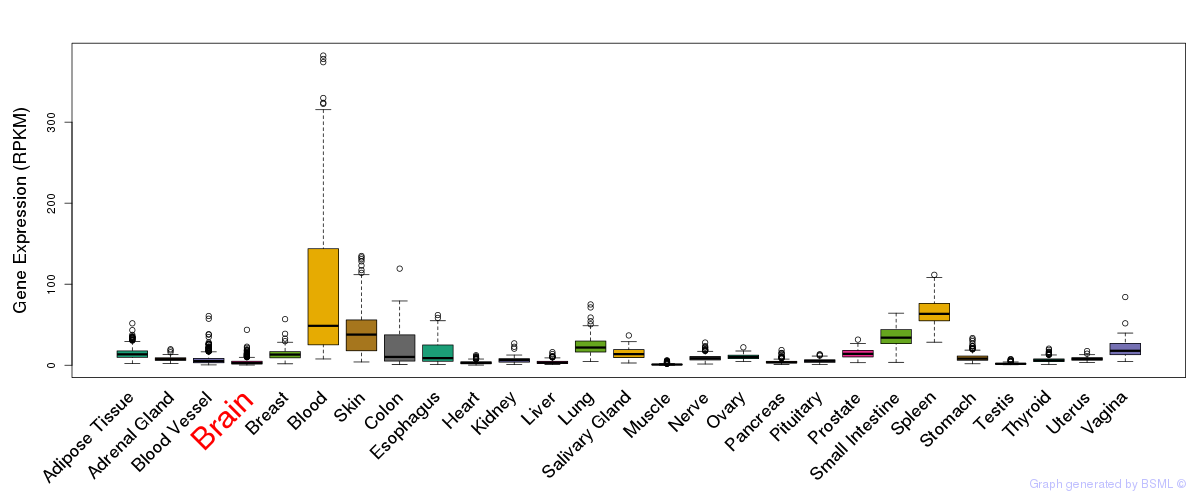
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 12019269 | |
| GO:0004197 | cysteine-type endopeptidase activity | IEA | - | |
| GO:0008656 | caspase activator activity | NAS | 12019269 | |
| GO:0032090 | Pyrin domain binding | IPI | 15030775 | |
| GO:0042803 | protein homodimerization activity | IDA | 15030775 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006508 | proteolysis | IEA | - | |
| GO:0006917 | induction of apoptosis | TAS | 10567338 | |
| GO:0007165 | signal transduction | NAS | 12019269 | |
| GO:0006954 | inflammatory response | IEA | - | |
| GO:0006919 | caspase activation | NAS | 12019269 | |
| GO:0033209 | tumor necrosis factor-mediated signaling pathway | IDA | 12656673 | |
| GO:0051092 | positive regulation of NF-kappaB transcription factor activity | IDA | 12656673 | |
| GO:0050718 | positive regulation of interleukin-1 beta secretion | IDA | 15030775 | |
| GO:0045786 | negative regulation of cell cycle | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | IEA | - | |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005737 | cytoplasm | NAS | 12019269 | |
| GO:0008385 | IkappaB kinase complex | TAS | 12656673 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NOD LIKE RECEPTOR SIGNALING PATHWAY | 62 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG CYTOSOLIC DNA SENSING PATHWAY | 56 | 44 | All SZGR 2.0 genes in this pathway |
| PID P53 DOWNSTREAM PATHWAY | 137 | 94 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME THE NLRP3 INFLAMMASOME | 12 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME INFLAMMASOMES | 17 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR NLR SIGNALING PATHWAYS | 46 | 33 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| DOANE RESPONSE TO ANDROGEN DN | 241 | 146 | All SZGR 2.0 genes in this pathway |
| NOJIMA SFRP2 TARGETS DN | 25 | 18 | All SZGR 2.0 genes in this pathway |
| JAEGER METASTASIS DN | 258 | 141 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 UP | 380 | 236 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 UP | 418 | 263 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| HUMMERICH SKIN CANCER PROGRESSION UP | 88 | 58 | All SZGR 2.0 genes in this pathway |
| FURUKAWA DUSP6 TARGETS PCI35 UP | 74 | 32 | All SZGR 2.0 genes in this pathway |
| SCHLESINGER METHYLATED DE NOVO IN CANCER | 88 | 64 | All SZGR 2.0 genes in this pathway |
| OHM METHYLATED IN ADULT CANCERS | 27 | 23 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| ROSS AML WITH MLL FUSIONS | 78 | 45 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS INTERMEDIATE PROGENITOR | 149 | 84 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| WANG LSD1 TARGETS UP | 24 | 14 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE DN | 258 | 160 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS DN | 306 | 191 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| PHESSE TARGETS OF APC AND MBD2 UP | 20 | 14 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| KIM GLIS2 TARGETS UP | 84 | 61 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| ZWANG EGF INTERVAL DN | 214 | 124 | All SZGR 2.0 genes in this pathway |