Gene Page: GRM4
Summary ?
| GeneID | 2914 |
| Symbol | GRM4 |
| Synonyms | GPRC1D|MGLUR4|mGlu4 |
| Description | glutamate receptor, metabotropic 4 |
| Reference | MIM:604100|HGNC:HGNC:4596|Ensembl:ENSG00000124493|HPRD:04978| |
| Gene type | protein-coding |
| Map location | 6p21.3 |
| Pascal p-value | 0.119 |
| Sherlock p-value | 0.722 |
| Fetal beta | -0.155 |
| eGene | Cerebellar Hemisphere Myers' cis & trans |
| Support | GPCR SIGNALLING CompositeSet Darnell FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 1 | Link to SZGene |
| GSMA_I | Genome scan meta-analysis | Psr: 0.033 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.04433 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 5 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0018 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17392743 | chr1 | 54764240 | GRM4 | 2914 | 0.01 | trans | ||
| rs6687849 | chr1 | 175904808 | GRM4 | 2914 | 0 | trans | ||
| rs2481653 | chr1 | 176044120 | GRM4 | 2914 | 0.05 | trans | ||
| rs2502827 | chr1 | 176044216 | GRM4 | 2914 | 1.129E-5 | trans | ||
| rs12405921 | chr1 | 206695730 | GRM4 | 2914 | 0.03 | trans | ||
| rs17572651 | chr1 | 218943612 | GRM4 | 2914 | 1.145E-4 | trans | ||
| snp_a-1919794 | 0 | GRM4 | 2914 | 0.19 | trans | |||
| rs882011 | chr2 | 22711668 | GRM4 | 2914 | 0.06 | trans | ||
| rs16829545 | chr2 | 151977407 | GRM4 | 2914 | 1.247E-5 | trans | ||
| rs3845734 | chr2 | 171125572 | GRM4 | 2914 | 2.083E-4 | trans | ||
| rs7584986 | chr2 | 184111432 | GRM4 | 2914 | 2.423E-9 | trans | ||
| rs6807632 | chr3 | 111395567 | GRM4 | 2914 | 0 | trans | ||
| rs7635430 | chr3 | 180152035 | GRM4 | 2914 | 0.02 | trans | ||
| rs6828371 | chr4 | 15757600 | GRM4 | 2914 | 0.12 | trans | ||
| rs6826731 | chr4 | 72130691 | GRM4 | 2914 | 0.06 | trans | ||
| rs2115110 | chr5 | 38193428 | GRM4 | 2914 | 0.12 | trans | ||
| rs17762315 | chr5 | 76807576 | GRM4 | 2914 | 0 | trans | ||
| rs7729096 | chr5 | 76835927 | GRM4 | 2914 | 0 | trans | ||
| rs10063752 | chr5 | 109259572 | GRM4 | 2914 | 0.08 | trans | ||
| rs1368303 | chr5 | 147672388 | GRM4 | 2914 | 1.145E-7 | trans | ||
| rs1929769 | chr6 | 40765977 | GRM4 | 2914 | 0.13 | trans | ||
| rs2530313 | chr7 | 148112233 | GRM4 | 2914 | 0.17 | trans | ||
| rs17079498 | chr8 | 3018343 | GRM4 | 2914 | 0.02 | trans | ||
| rs4872540 | chr8 | 22598980 | GRM4 | 2914 | 0.05 | trans | ||
| rs6989594 | chr8 | 126303866 | GRM4 | 2914 | 0.02 | trans | ||
| rs7021724 | chr9 | 2527899 | GRM4 | 2914 | 0.13 | trans | ||
| rs2766987 | chr9 | 113667956 | GRM4 | 2914 | 0.12 | trans | ||
| rs17263352 | chr9 | 124811572 | GRM4 | 2914 | 0.02 | trans | ||
| rs12716680 | chr13 | 42196469 | GRM4 | 2914 | 0.12 | trans | ||
| rs7996604 | chr13 | 42224412 | GRM4 | 2914 | 0.07 | trans | ||
| snp_a-1880548 | 0 | GRM4 | 2914 | 0.08 | trans | |||
| rs1629937 | chr14 | 77059671 | GRM4 | 2914 | 0 | trans | ||
| rs17104720 | chr14 | 77127308 | GRM4 | 2914 | 2.101E-6 | trans | ||
| rs17104722 | chr14 | 77138109 | GRM4 | 2914 | 0.07 | trans | ||
| rs17104782 | chr14 | 77156735 | GRM4 | 2914 | 0.07 | trans | ||
| rs6574467 | chr14 | 79179744 | GRM4 | 2914 | 0.01 | trans | ||
| rs10146003 | chr14 | 79191170 | GRM4 | 2914 | 0.01 | trans | ||
| rs16955618 | chr15 | 29937543 | GRM4 | 2914 | 2.74E-23 | trans | ||
| snp_a-1830894 | 0 | GRM4 | 2914 | 6.868E-4 | trans | |||
| rs16968252 | chr17 | 31692447 | GRM4 | 2914 | 0.08 | trans | ||
| rs17823633 | chr17 | 68851458 | GRM4 | 2914 | 0.15 | trans | ||
| rs17085767 | chr18 | 69839397 | GRM4 | 2914 | 0.1 | trans | ||
| rs749043 | chr20 | 46995700 | GRM4 | 2914 | 0.2 | trans | ||
| rs1041786 | chr21 | 22617710 | GRM4 | 2914 | 0.19 | trans |
Section II. Transcriptome annotation
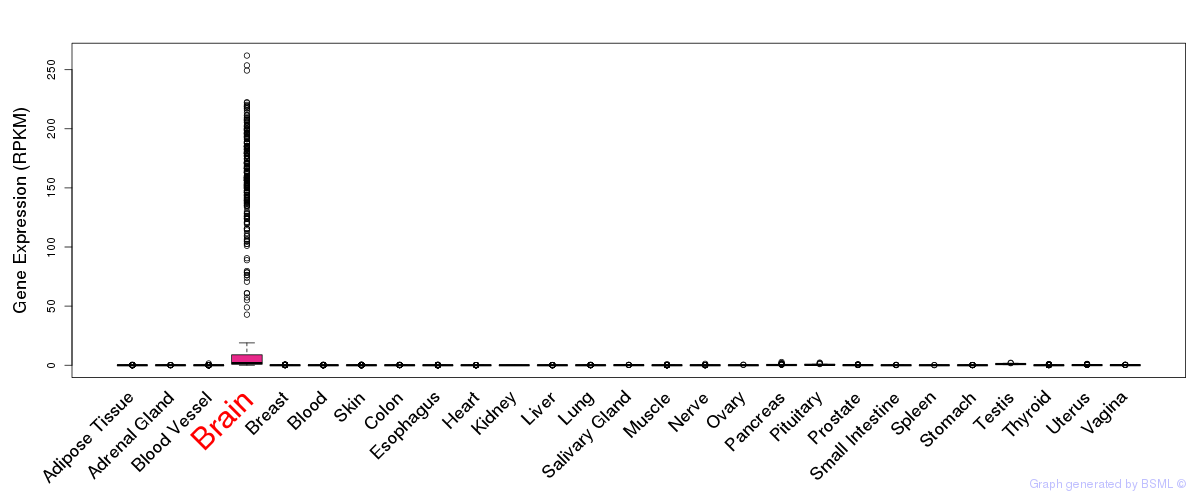
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TBC1D25 | 0.92 | 0.91 |
| APBB1 | 0.91 | 0.90 |
| AGAP3 | 0.91 | 0.89 |
| AP2A2 | 0.91 | 0.91 |
| EPN1 | 0.90 | 0.89 |
| RTN1 | 0.90 | 0.88 |
| CACNB3 | 0.89 | 0.89 |
| SNAP47 | 0.89 | 0.84 |
| MCOLN1 | 0.89 | 0.88 |
| ASB6 | 0.89 | 0.89 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.78 | -0.71 |
| MT-CO2 | -0.77 | -0.66 |
| AF347015.31 | -0.77 | -0.66 |
| AF347015.8 | -0.77 | -0.67 |
| MT-CYB | -0.75 | -0.65 |
| AF347015.2 | -0.74 | -0.61 |
| AF347015.26 | -0.74 | -0.63 |
| AF347015.33 | -0.74 | -0.62 |
| AF347015.27 | -0.73 | -0.66 |
| AL139819.3 | -0.72 | -0.69 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004872 | receptor activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007269 | neurotransmitter secretion | TAS | neuron, axon, Synap, serotonin, Neurotransmitter, dopamine (GO term level: 8) | 15102938 |
| GO:0007268 | synaptic transmission | IEA | neuron, Synap, Neurotransmitter (GO term level: 6) | - |
| GO:0007268 | synaptic transmission | TAS | neuron, Synap, Neurotransmitter (GO term level: 6) | 8738157 |
| GO:0007196 | metabotropic glutamate receptor, adenylate cyclase inhibiting pathway | IMP | glutamate (GO term level: 12) | 15102938 |
| GO:0043526 | neuroprotection | TAS | neuron (GO term level: 9) | 15102938 |
| GO:0000187 | activation of MAPK activity | IDA | 15102938 | |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | IEA | - | |
| GO:0007194 | negative regulation of adenylate cyclase activity | IEP | 15102938 | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0007612 | learning | IEA | - | |
| GO:0043410 | positive regulation of MAPKKK cascade | IEP | 15102938 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005622 | intracellular | IDA | 15102938 | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | IDA | 15102938 | |
| GO:0031410 | cytoplasmic vesicle | IDA | 15102938 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| KEGG TASTE TRANSDUCTION | 52 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | 15 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-129-5p | 668 | 674 | 1A | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-203.1 | 937 | 943 | m8 | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-29 | 122 | 128 | m8 | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.