Gene Page: GTF2E1
Summary ?
| GeneID | 2960 |
| Symbol | GTF2E1 |
| Synonyms | FE|TF2E1|TFIIE-A |
| Description | general transcription factor IIE subunit 1 |
| Reference | MIM:189962|HGNC:HGNC:4650|Ensembl:ENSG00000153767|HPRD:01799|Vega:OTTHUMG00000159667 |
| Gene type | protein-coding |
| Map location | 3q21-q24 |
| Pascal p-value | 0.599 |
| DEG p-value | DEG:Zhao_2015:p=5.81e-04:q=0.0926 |
| Fetal beta | 0.111 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DEG:Zhao_2015 | RNA Sequencing analysis | Transcriptome sequencing and genome-wide association analyses reveal lysosomal function and actin cytoskeleton remodeling in schizophrenia and bipolar disorder. |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
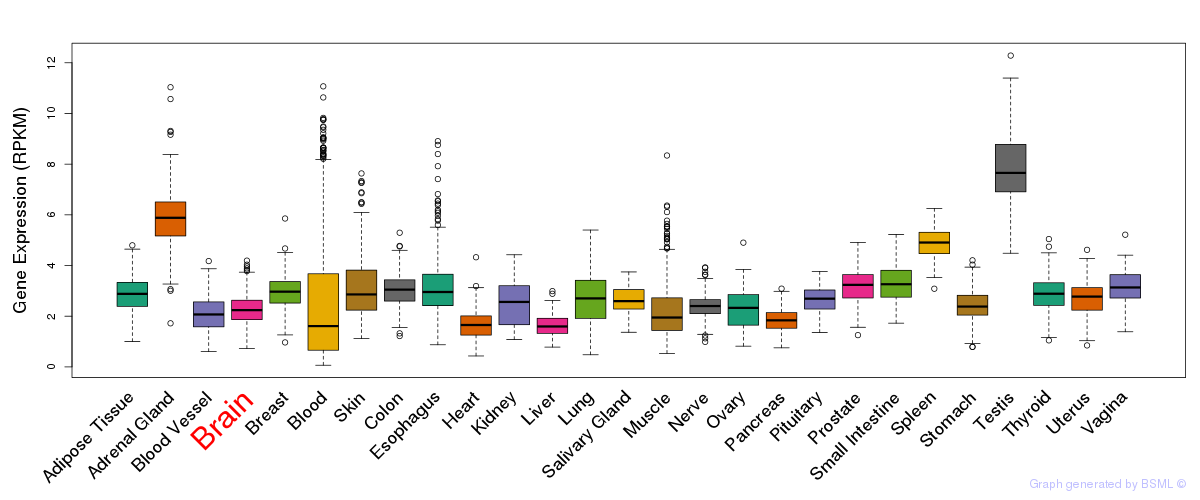
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PPM1L | 0.91 | 0.92 |
| GPD1L | 0.90 | 0.91 |
| MMP16 | 0.87 | 0.89 |
| C1orf96 | 0.87 | 0.92 |
| BMPR2 | 0.86 | 0.92 |
| NLGN1 | 0.86 | 0.90 |
| LASS6 | 0.86 | 0.89 |
| LRRTM2 | 0.85 | 0.89 |
| DNM3 | 0.85 | 0.89 |
| PDE4D | 0.85 | 0.89 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SERPINB6 | -0.63 | -0.71 |
| C19orf36 | -0.60 | -0.69 |
| HSD17B14 | -0.60 | -0.73 |
| FXYD1 | -0.59 | -0.77 |
| FAH | -0.59 | -0.69 |
| HEPN1 | -0.58 | -0.70 |
| TREX1 | -0.58 | -0.70 |
| TTYH1 | -0.58 | -0.66 |
| ROM1 | -0.57 | -0.72 |
| TLCD1 | -0.57 | -0.74 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AES | AES-1 | AES-2 | ESP1 | GRG | GRG5 | TLE5 | amino-terminal enhancer of split | - | HPRD,BioGRID | 11416139 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | Co-purification | BioGRID | 9159119 |
| ERCC3 | BTF2 | GTF2H | RAD25 | TFIIH | XPB | excision repair cross-complementing rodent repair deficiency, complementation group 3 (xeroderma pigmentosum group B complementing) | - | HPRD,BioGRID | 7926747 |8152490 |
| GTF2A1 | MGC129969 | MGC129970 | TF2A1 | TFIIA | general transcription factor IIA, 1, 19/37kDa | - | HPRD,BioGRID | 11509574 |
| GTF2A2 | HsT18745 | TF2A2 | TFIIA | general transcription factor IIA, 2, 12kDa | - | HPRD | 11509574 |
| GTF2B | TF2B | TFIIB | general transcription factor IIB | Co-purification Reconstituted Complex | BioGRID | 9159119 |
| GTF2E2 | FE | TF2E2 | TFIIE-B | general transcription factor IIE, polypeptide 2, beta 34kDa | - | HPRD,BioGRID | 9677423 |11113176 |12665589 |
| GTF2F1 | BTF4 | RAP74 | TF2F1 | TFIIF | general transcription factor IIF, polypeptide 1, 74kDa | Co-purification Reconstituted Complex | BioGRID | 7926747 |9159119 |
| GTF2F2 | BTF4 | RAP30 | TF2F2 | TFIIF | general transcription factor IIF, polypeptide 2, 30kDa | - | HPRD | 11113176 |
| GTF2F2 | BTF4 | RAP30 | TF2F2 | TFIIF | general transcription factor IIF, polypeptide 2, 30kDa | Reconstituted Complex | BioGRID | 7926747 |
| GTF2H1 | BTF2 | TFB1 | TFIIH | general transcription factor IIH, polypeptide 1, 62kDa | - | HPRD | 11113176 |
| GTF2H4 | TFB2 | TFIIH | general transcription factor IIH, polypeptide 4, 52kDa | Co-purification Reconstituted Complex | BioGRID | 9159119 |
| GTF2H4 | TFB2 | TFIIH | general transcription factor IIH, polypeptide 4, 52kDa | - | HPRD | 11113176 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | Reconstituted Complex | BioGRID | 9271120 |
| MED21 | SRB7 | SURB7 | mediator complex subunit 21 | Affinity Capture-Western Co-purification | BioGRID | 9159119 |
| POLR2A | MGC75453 | POLR2 | POLRA | RPB1 | RPBh1 | RPO2 | RPOL2 | RpIILS | hRPB220 | hsRPB1 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | Co-purification Reconstituted Complex | BioGRID | 7926747 |9159119 |
| SND1 | TDRD11 | p100 | staphylococcal nuclease and tudor domain containing 1 | - | HPRD,BioGRID | 7651391 |
| TAF1 | BA2R | CCG1 | CCGS | DYT3 | KAT4 | N-TAF1 | NSCL2 | OF | P250 | TAF2A | TAFII250 | TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa | Reconstituted Complex | BioGRID | 7926747 |
| TAF11 | MGC:15243 | PRO2134 | TAF2I | TAFII28 | TAF11 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 28kDa | Co-purification Reconstituted Complex | BioGRID | 9159119 |
| TAF6 | DKFZp781E21155 | MGC:8964 | TAF2E | TAFII70 | TAFII80 | TAFII85 | TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa | - | HPRD,BioGRID | 7667268 |
| TBP | GTF2D | GTF2D1 | MGC117320 | MGC126054 | MGC126055 | SCA17 | TFIID | TATA box binding protein | - | HPRD | 11113176 |
| TBP | GTF2D | GTF2D1 | MGC117320 | MGC126054 | MGC126055 | SCA17 | TFIID | TATA box binding protein | Co-purification Reconstituted Complex | BioGRID | 7926747 |9159119 |
| TCEA1 | GTF2S | SII | TCEA | TF2S | TFIIS | transcription elongation factor A (SII), 1 | - | HPRD,BioGRID | 9305922 |
| TRIM24 | PTC6 | RNF82 | TF1A | TIF1 | TIF1A | TIF1ALPHA | hTIF1 | tripartite motif-containing 24 | - | HPRD,BioGRID | 9632676 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG BASAL TRANSCRIPTION FACTORS | 36 | 24 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CARM ER PATHWAY | 35 | 27 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RARRXR PATHWAY | 15 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL II TRANSCRIPTION | 105 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | 41 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTION | 210 | 127 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | 61 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV INFECTION | 207 | 122 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV LIFE CYCLE | 125 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME LATE PHASE OF HIV LIFE CYCLE | 104 | 61 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 UP | 209 | 139 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA HIF1A UP | 77 | 49 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| SHIPP DLBCL CURED VS FATAL DN | 45 | 30 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND DN | 225 | 163 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D2 | 41 | 30 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| RIGGINS TAMOXIFEN RESISTANCE DN | 220 | 147 | All SZGR 2.0 genes in this pathway |
| KAMMINGA SENESCENCE | 41 | 26 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 1HR DN | 222 | 147 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR DN | 254 | 158 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |