Gene Page: EPN1
Summary ?
| GeneID | 29924 |
| Symbol | EPN1 |
| Synonyms | - |
| Description | epsin 1 |
| Reference | MIM:607262|HGNC:HGNC:21604|Ensembl:ENSG00000063245|HPRD:06270|Vega:OTTHUMG00000180911 |
| Gene type | protein-coding |
| Map location | 19q13.42 |
| Pascal p-value | 0.058 |
| Sherlock p-value | 0.233 |
| Fetal beta | -0.313 |
| DMG | 1 (# studies) |
| Support | CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 1 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0058 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg17387916 | 19 | 55896965 | EPN1 | 3.549E-4 | 3.328 | DMG:vanEijk_2014 |
Section II. Transcriptome annotation
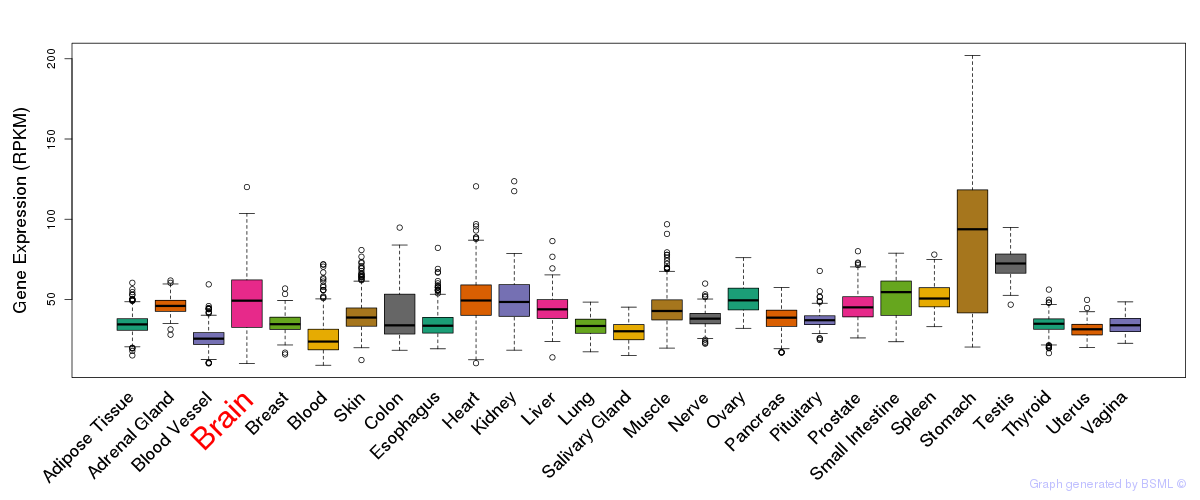
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0008289 | lipid binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006897 | endocytosis | TAS | 9723620 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005905 | coated pit | IEA | - | |
| GO:0005886 | plasma membrane | EXP | 10567358 |12218189 |15962011 | |
| GO:0005886 | plasma membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AP2A1 | ADTAA | AP2-ALPHA | CLAPA1 | adaptor-related protein complex 2, alpha 1 subunit | - | HPRD | 9723620 |
| AP2A2 | ADTAB | CLAPA2 | HIP9 | HYPJ | adaptor-related protein complex 2, alpha 2 subunit | Affinity Capture-Western Reconstituted Complex | BioGRID | 9723620 |
| CLTC | CHC | CHC17 | CLH-17 | CLTCL2 | Hc | KIAA0034 | clathrin, heavy chain (Hc) | - | HPRD,BioGRID | 10692452 |
| CRMP1 | DPYSL1 | DRP-1 | DRP1 | collapsin response mediator protein 1 | Two-hybrid | BioGRID | 16169070 |
| EPS15 | AF-1P | AF1P | MLLT5 | epidermal growth factor receptor pathway substrate 15 | - | HPRD,BioGRID | 9723620 |
| EPS15L1 | EPS15R | epidermal growth factor receptor pathway substrate 15-like 1 | Affinity Capture-Western | BioGRID | 9723620 |
| GTF3A | AP2 | TFIIIA | general transcription factor IIIA | Affinity Capture-Western | BioGRID | 12775724 |
| KIAA1377 | - | KIAA1377 | Two-hybrid | BioGRID | 16169070 |
| MED31 | 3110004H13Rik | CGI-125 | FLJ27436 | FLJ36714 | Soh1 | mediator complex subunit 31 | Two-hybrid | BioGRID | 16169070 |
| RALBP1 | RIP1 | RLIP1 | RLIP76 | ralA binding protein 1 | Affinity Capture-Western | BioGRID | 12775724 |
| REPS2 | POB1 | RALBP1 associated Eps domain containing 2 | - | HPRD,BioGRID | 10557078 |
| ZBTB16 | PLZF | ZNF145 | zinc finger and BTB domain containing 16 | - | HPRD | 10791968 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ENDOCYTOSIS | 183 | 132 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NDKDYNAMIN PATHWAY | 21 | 15 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 INTERNALIZATION PATHWAY | 41 | 35 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY EGFR IN CANCER | 109 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME EGFR DOWNREGULATION | 25 | 14 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE DN | 244 | 147 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER UP | 298 | 174 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-23 | 75 | 81 | 1A | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.