Gene Page: UBQLN1
Summary ?
| GeneID | 29979 |
| Symbol | UBQLN1 |
| Synonyms | DA41|DSK2|PLIC-1|UBQN|XDRP1 |
| Description | ubiquilin 1 |
| Reference | MIM:605046|HGNC:HGNC:12508|Ensembl:ENSG00000135018|HPRD:05440|Vega:OTTHUMG00000020104 |
| Gene type | protein-coding |
| Map location | 9q22|9q21.2-q21.3 |
| Pascal p-value | 0.028 |
| Sherlock p-value | 0.259 |
| TADA p-value | 0.011 |
| Fetal beta | 0.455 |
| Support | CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Xu_2012 | Whole Exome Sequencing analysis | De novo mutations of 4 genes were identified by exome sequencing of 795 samples in this study | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| UBQLN1 | chr9 | 86292782 | T | A | NM_013438 NM_053067 | p.322N>I p.322N>I | missense missense | Schizophrenia | DNM:Xu_2012 |
Section II. Transcriptome annotation
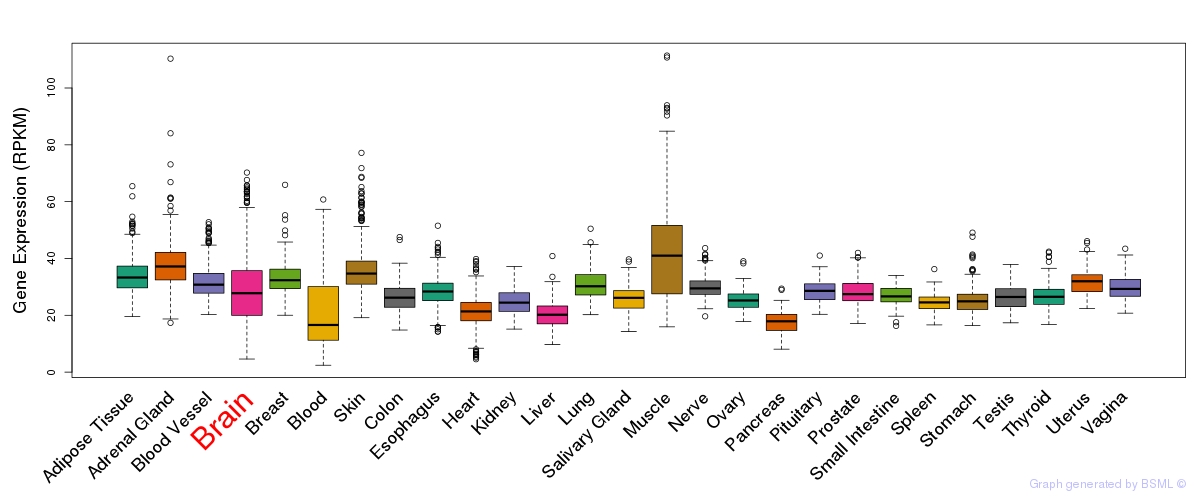
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CD47 | IAP | MER6 | OA3 | CD47 molecule | - | HPRD | 10549293 |
| CD99 | MIC2 | MIC2X | MIC2Y | CD99 molecule | Two-hybrid | BioGRID | 16189514 |
| CSTF2 | CstF-64 | cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa | Two-hybrid | BioGRID | 16189514 |
| CSTF2T | CstF-64T | DKFZp434C1013 | KIAA0689 | cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa, tau variant | Two-hybrid | BioGRID | 16189514 |
| EFEMP2 | FBLN4 | MBP1 | UPH1 | EGF-containing fibulin-like extracellular matrix protein 2 | Two-hybrid | BioGRID | 16189514 |
| FAM127B | CXX1b | DKFZp564B147 | MAR8A | MGC8471 | family with sequence similarity 127, member B | Two-hybrid | BioGRID | 16189514 |
| FRAP1 | FLJ44809 | FRAP | FRAP2 | MTOR | RAFT1 | RAPT1 | FK506 binding protein 12-rapamycin associated protein 1 | - | HPRD,BioGRID | 11853878 |
| FZD7 | FzE3 | frizzled homolog 7 (Drosophila) | Two-hybrid | BioGRID | 16189514 |
| GABRD | MGC45284 | gamma-aminobutyric acid (GABA) A receptor, delta | Two-hybrid | BioGRID | 16189514 |
| HGS | HRS | ZFYVE8 | hepatocyte growth factor-regulated tyrosine kinase substrate | Two-hybrid | BioGRID | 16189514 |
| HK2 | DKFZp686M1669 | HKII | HXK2 | hexokinase 2 | Two-hybrid | BioGRID | 16189514 |
| HSPA13 | STCH | heat shock protein 70kDa family, member 13 | Two-hybrid | BioGRID | 16189514 |
| IGLC1 | IGLC | immunoglobulin lambda constant 1 (Mcg marker) | Two-hybrid | BioGRID | 16189514 |
| KLHDC5 | KIAA1340 | MGC131714 | kelch domain containing 5 | Two-hybrid | BioGRID | 16189514 |
| NBL1 | D1S1733E | DAN | DAND1 | MGC8972 | NB | NO3 | neuroblastoma, suppression of tumorigenicity 1 | - | HPRD,BioGRID | 9303440 |10390159 |
| P4HB | DSI | ERBA2L | GIT | PDI | PDIA1 | PHDB | PO4DB | PO4HB | PROHB | procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), beta polypeptide | - | HPRD,BioGRID | 12095988 |
| PLA2G16 | AdPLA | H-REV107-1 | HRASLS3 | HREV107 | HREV107-3 | MGC118754 | phospholipase A2, group XVI | Two-hybrid | BioGRID | 16189514 |
| PSEN1 | AD3 | FAD | PS1 | S182 | presenilin 1 | Reconstituted Complex Two-hybrid | BioGRID | 11076969 |
| PSEN2 | AD3L | AD4 | PS2 | STM2 | presenilin 2 (Alzheimer disease 4) | - | HPRD,BioGRID | 11076969 |
| RIC8A | MGC104517 | MGC131931 | MGC148073 | MGC148074 | RIC8 | synembryn | resistance to inhibitors of cholinesterase 8 homolog A (C. elegans) | Two-hybrid | BioGRID | 16189514 |
| SERPINI2 | MEPI | PANCPIN | PI14 | TSA2004 | serpin peptidase inhibitor, clade I (pancpin), member 2 | Two-hybrid | BioGRID | 16189514 |
| SMCR7 | MGC23130 | Smith-Magenis syndrome chromosome region, candidate 7 | Two-hybrid | BioGRID | 16189514 |
| TFF1 | BCEI | D21S21 | HP1.A | HPS2 | pNR-2 | pS2 | trefoil factor 1 | Two-hybrid | BioGRID | 16189514 |
| TMCO6 | FLJ39769 | PRO1580 | transmembrane and coiled-coil domains 6 | Two-hybrid | BioGRID | 16189514 |
| TRIM32 | BBS11 | HT2A | LGMD2H | TATIP | tripartite motif-containing 32 | Two-hybrid | BioGRID | 16189514 |
| UBQLN1 | DA41 | DSK2 | FLJ90054 | PLIC-1 | XDRP1 | ubiquilin 1 | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID CXCR4 PATHWAY | 102 | 78 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS DN | 352 | 225 | All SZGR 2.0 genes in this pathway |
| WANG CLIM2 TARGETS DN | 186 | 114 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2A DN | 141 | 84 | All SZGR 2.0 genes in this pathway |
| APPIERTO RESPONSE TO FENRETINIDE UP | 38 | 26 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| GEISS RESPONSE TO DSRNA UP | 38 | 29 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE UP | 149 | 85 | All SZGR 2.0 genes in this pathway |
| COLINA TARGETS OF 4EBP1 AND 4EBP2 | 356 | 214 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE UP | 442 | 263 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 11 | 103 | 68 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |