Gene Page: HAL
Summary ?
| GeneID | 3034 |
| Symbol | HAL |
| Synonyms | HIS|HSTD |
| Description | histidine ammonia-lyase |
| Reference | MIM:609457|HGNC:HGNC:4806|Ensembl:ENSG00000084110|HPRD:07187|Vega:OTTHUMG00000170354 |
| Gene type | protein-coding |
| Map location | 12q23.1 |
| Pascal p-value | 0.94 |
| Fetal beta | 0.32 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 2 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg04488758 | 12 | 95945366 | HAL | 0.004 | 4.129 | DMG:vanEijk_2014 | |
| cg27246571 | 12 | 96389588 | HAL | 0.004 | -4.064 | DMG:vanEijk_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs695809 | chr22 | 26278127 | HAL | 3034 | 0.11 | trans |
Section II. Transcriptome annotation
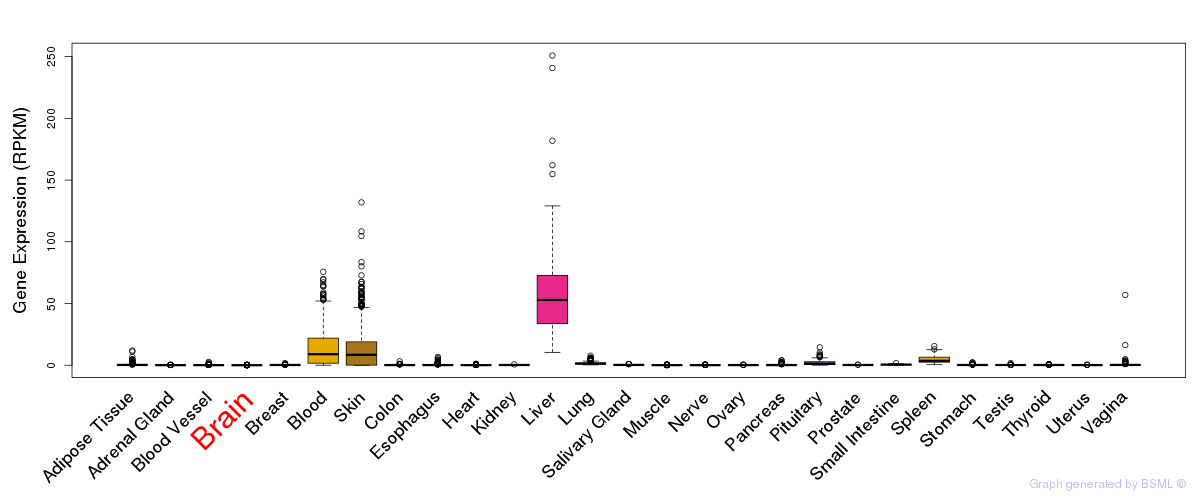
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG HISTIDINE METABOLISM | 29 | 19 | All SZGR 2.0 genes in this pathway |
| KEGG NITROGEN METABOLISM | 23 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | 200 | 136 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA UP | 177 | 110 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA DN | 116 | 79 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL DN | 244 | 153 | All SZGR 2.0 genes in this pathway |
| JAEGER METASTASIS DN | 258 | 141 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA UP | 544 | 308 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LPS UP | 431 | 237 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 3 4WK UP | 214 | 144 | All SZGR 2.0 genes in this pathway |
| HOWLIN PUBERTAL MAMMARY GLAND | 69 | 40 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER CIPROFIBRATE DN | 66 | 43 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC TGFA DN | 65 | 44 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER ACOX1 DN | 65 | 39 | All SZGR 2.0 genes in this pathway |
| UEDA PERIFERAL CLOCK | 169 | 111 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC E2F1 DN | 64 | 38 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC DN | 63 | 40 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER E2F1 DN | 64 | 42 | All SZGR 2.0 genes in this pathway |
| HSIAO LIVER SPECIFIC GENES | 244 | 154 | All SZGR 2.0 genes in this pathway |
| HADDAD T LYMPHOCYTE AND NK PROGENITOR DN | 63 | 41 | All SZGR 2.0 genes in this pathway |
| HOWLIN CITED1 TARGETS 1 UP | 35 | 25 | All SZGR 2.0 genes in this pathway |
| CADWELL ATG16L1 TARGETS DN | 70 | 43 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS CTNNB1 DN | 170 | 105 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA DN | 267 | 160 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| KINNEY DNMT1 METHYLATION TARGETS | 14 | 6 | All SZGR 2.0 genes in this pathway |