Gene Page: SERPIND1
Summary ?
| GeneID | 3053 |
| Symbol | SERPIND1 |
| Synonyms | D22S673|HC2|HCF2|HCII|HLS2|LS2|THPH10 |
| Description | serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| Reference | MIM:142360|HGNC:HGNC:4838|Ensembl:ENSG00000099937|HPRD:00795|Vega:OTTHUMG00000150755 |
| Gene type | protein-coding |
| Map location | 22q11.21 |
| Pascal p-value | 0.252 |
| Fetal beta | 0.567 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CNV:YES | Copy number variation studies | Manual curation | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.031 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17131472 | chr1 | 91962966 | SERPIND1 | 3053 | 0.09 | trans | ||
| rs6718379 | chr2 | 18632808 | SERPIND1 | 3053 | 0.13 | trans | ||
| rs17015803 | chr2 | 120191430 | SERPIND1 | 3053 | 0.03 | trans | ||
| rs2922180 | chr3 | 125280472 | SERPIND1 | 3053 | 0.03 | trans | ||
| rs2971271 | chr3 | 125450402 | SERPIND1 | 3053 | 0.14 | trans | ||
| rs2976809 | chr3 | 125451738 | SERPIND1 | 3053 | 1.705E-4 | trans | ||
| rs6449169 | 0 | SERPIND1 | 3053 | 0.18 | trans | |||
| rs313946 | chr4 | 113975904 | SERPIND1 | 3053 | 0.07 | trans | ||
| rs657141 | chr4 | 113981041 | SERPIND1 | 3053 | 0.16 | trans | ||
| rs17381134 | chr4 | 157179855 | SERPIND1 | 3053 | 0.08 | trans | ||
| rs2294262 | chr6 | 16108966 | SERPIND1 | 3053 | 0.19 | trans | ||
| rs4236095 | chr6 | 47368686 | SERPIND1 | 3053 | 0.16 | trans | ||
| rs6455226 | chr6 | 67930295 | SERPIND1 | 3053 | 0.01 | trans | ||
| rs2718035 | chr7 | 36132888 | SERPIND1 | 3053 | 0.03 | trans | ||
| rs3110902 | chr7 | 65375101 | SERPIND1 | 3053 | 0.13 | trans | ||
| rs7841407 | chr8 | 9243427 | SERPIND1 | 3053 | 0.09 | trans | ||
| rs680372 | chr8 | 36999626 | SERPIND1 | 3053 | 0.18 | trans | ||
| rs4514358 | chr10 | 43861923 | SERPIND1 | 3053 | 0.02 | trans | ||
| rs11016704 | chr10 | 128989145 | SERPIND1 | 3053 | 0.07 | trans | ||
| rs17134872 | chr11 | 76139845 | SERPIND1 | 3053 | 0.16 | trans | ||
| rs11236774 | chr11 | 76234952 | SERPIND1 | 3053 | 0.16 | trans | ||
| rs9529974 | chr13 | 72821935 | SERPIND1 | 3053 | 0 | trans | ||
| rs7164640 | chr15 | 80661303 | SERPIND1 | 3053 | 0.12 | trans | ||
| rs11873703 | chr18 | 61448702 | SERPIND1 | 3053 | 0.03 | trans | ||
| snp_a-1913345 | 0 | SERPIND1 | 3053 | 0.12 | trans | |||
| rs293737 | chr20 | 31925477 | SERPIND1 | 3053 | 0.1 | trans | ||
| rs6095741 | chr20 | 48666589 | SERPIND1 | 3053 | 0 | trans | ||
| rs17255719 | chrX | 12144766 | SERPIND1 | 3053 | 0.09 | trans | ||
| rs5991662 | chrX | 43335796 | SERPIND1 | 3053 | 0.06 | trans | ||
| rs6529399 | chrX | 128763582 | SERPIND1 | 3053 | 0.04 | trans | ||
| rs3761581 | chrX | 128789720 | SERPIND1 | 3053 | 0 | trans | ||
| rs242163 | chrX | 132311662 | SERPIND1 | 3053 | 0.02 | trans |
Section II. Transcriptome annotation
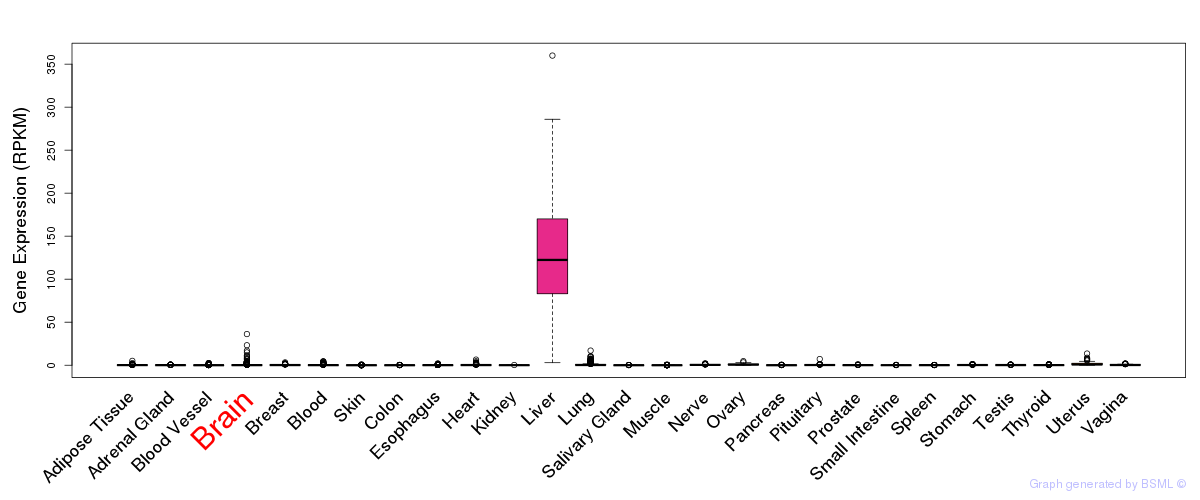
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BRIP1 | 0.97 | 0.76 |
| FANCI | 0.97 | 0.70 |
| MELK | 0.97 | 0.83 |
| KIF23 | 0.97 | 0.75 |
| SMC4 | 0.96 | 0.79 |
| BUB1B | 0.96 | 0.79 |
| ERCC6L | 0.96 | 0.73 |
| BRCA2 | 0.96 | 0.80 |
| BUB1 | 0.96 | 0.79 |
| MCM10 | 0.96 | 0.79 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FBXO2 | -0.45 | -0.70 |
| SLC9A3R2 | -0.45 | -0.54 |
| LHPP | -0.45 | -0.66 |
| HLA-F | -0.45 | -0.73 |
| PTH1R | -0.44 | -0.69 |
| C5orf53 | -0.44 | -0.68 |
| ASPHD1 | -0.44 | -0.62 |
| TNFSF12 | -0.44 | -0.66 |
| ALDOC | -0.43 | -0.67 |
| AIFM3 | -0.43 | -0.68 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004867 | serine-type endopeptidase inhibitor activity | IEA | - | |
| GO:0008201 | heparin binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007596 | blood coagulation | IEA | - | |
| GO:0006935 | chemotaxis | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | NAS | 14718574 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG COMPLEMENT AND COAGULATION CASCADES | 69 | 49 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| HUMMERICH BENIGN SKIN TUMOR UP | 18 | 8 | All SZGR 2.0 genes in this pathway |
| MUELLER METHYLATED IN GLIOBLASTOMA | 40 | 22 | All SZGR 2.0 genes in this pathway |
| LE EGR2 TARGETS DN | 108 | 84 | All SZGR 2.0 genes in this pathway |
| KENNY CTNNB1 TARGETS UP | 50 | 30 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL AND BRAIN QTL TRANS | 185 | 114 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| HSIAO LIVER SPECIFIC GENES | 244 | 154 | All SZGR 2.0 genes in this pathway |
| MCDOWELL ACUTE LUNG INJURY UP | 45 | 29 | All SZGR 2.0 genes in this pathway |
| SU LIVER | 55 | 32 | All SZGR 2.0 genes in this pathway |
| WILSON PROTEASES AT TUMOR BONE INTERFACE UP | 21 | 14 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER SURVIVAL UP | 185 | 112 | All SZGR 2.0 genes in this pathway |
| GU PDEF TARGETS UP | 71 | 49 | All SZGR 2.0 genes in this pathway |
| COULOUARN TEMPORAL TGFB1 SIGNATURE DN | 138 | 99 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT DN | 222 | 141 | All SZGR 2.0 genes in this pathway |
| CERIBELLI PROMOTERS INACTIVE AND BOUND BY NFY | 44 | 20 | All SZGR 2.0 genes in this pathway |
| NABA ECM REGULATORS | 238 | 125 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |