Gene Page: KCNIP3
Summary ?
| GeneID | 30818 |
| Symbol | KCNIP3 |
| Synonyms | CSEN|DREAM|KCHIP3 |
| Description | potassium voltage-gated channel interacting protein 3 |
| Reference | MIM:604662|HGNC:HGNC:15523|Ensembl:ENSG00000115041|HPRD:05232|Vega:OTTHUMG00000130392 |
| Gene type | protein-coding |
| Map location | 2q21.1 |
| Pascal p-value | 0.228 |
| Sherlock p-value | 0.795 |
| Fetal beta | -1.997 |
| DMG | 1 (# studies) |
| eGene | Cerebellar Hemisphere Cerebellum Frontal Cortex BA9 Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0004 | |
| Expression | Meta-analysis of gene expression | P value: 1.492 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg07166291 | 2 | 95971994 | KCNIP3 | 2.854E-4 | 0.539 | 0.039 | DMG:Wockner_2014 |
| cg21350153 | 2 | 95982371 | KCNIP3 | 2.864E-4 | 0.256 | 0.039 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4654352 | chr1 | 29789342 | KCNIP3 | 30818 | 0.08 | trans |
Section II. Transcriptome annotation
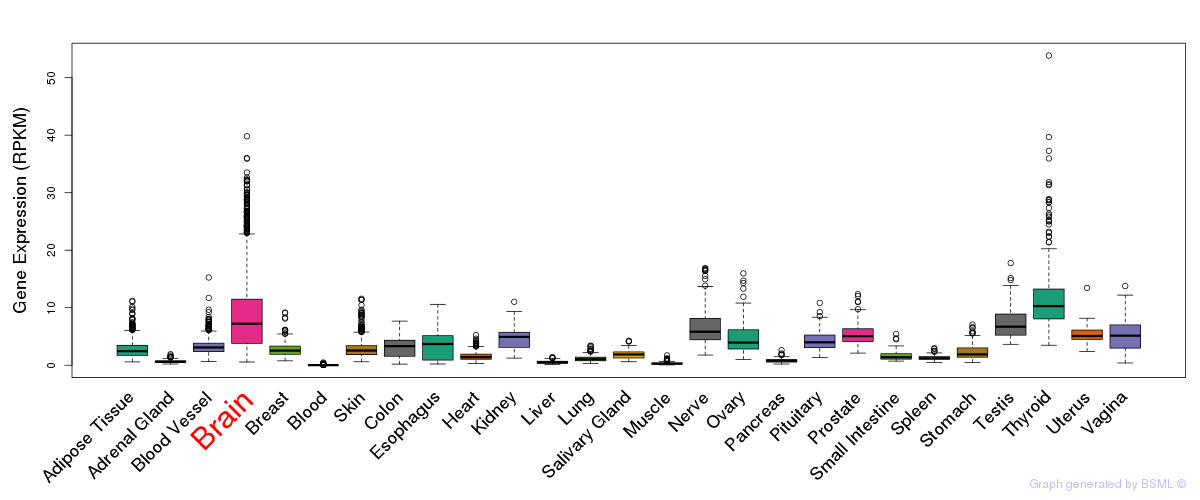
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KIAA2013 | 0.81 | 0.81 |
| RNH1 | 0.79 | 0.80 |
| C3orf39 | 0.79 | 0.80 |
| SCYL1 | 0.78 | 0.78 |
| GRINA | 0.77 | 0.77 |
| LRRC14 | 0.77 | 0.75 |
| DOHH | 0.77 | 0.78 |
| ZNF444 | 0.76 | 0.81 |
| C11orf68 | 0.76 | 0.76 |
| ATG4D | 0.76 | 0.77 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.46 | -0.38 |
| NSBP1 | -0.43 | -0.44 |
| AL139819.3 | -0.43 | -0.43 |
| EIF5B | -0.41 | -0.45 |
| AC010300.1 | -0.41 | -0.48 |
| AC090186.1 | -0.41 | -0.39 |
| MT-ATP8 | -0.41 | -0.34 |
| AC005921.3 | -0.40 | -0.44 |
| FAM159B | -0.40 | -0.57 |
| SYCP3 | -0.39 | -0.43 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003677 | DNA binding | TAS | 10078534 | |
| GO:0003714 | transcription corepressor activity | TAS | 10078534 | |
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0005509 | calcium ion binding | TAS | 10900016 | |
| GO:0005244 | voltage-gated ion channel activity | IEA | - | |
| GO:0005267 | potassium channel activity | IEA | - | |
| GO:0005216 | ion channel activity | IEA | - | |
| GO:0030955 | potassium ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006357 | regulation of transcription from RNA polymerase II promoter | TAS | 10078534 | |
| GO:0006350 | transcription | IEA | - | |
| GO:0007165 | signal transduction | TAS | 10900016 | |
| GO:0006811 | ion transport | IEA | - | |
| GO:0006813 | potassium ion transport | IEA | - | |
| GO:0006915 | apoptosis | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005794 | Golgi apparatus | IEA | - | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005783 | endoplasmic reticulum | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA DREAM PATHWAY | 14 | 13 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 LCP WITH H3K4ME3 | 162 | 80 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF LCP WITH H3K27ME3 | 70 | 35 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS LCP WITH H3K4ME3 | 174 | 100 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES LCP WITH H3K4ME3 | 142 | 80 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC LCP WITH H3K4ME3 | 58 | 34 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS UP | 387 | 225 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY LUMINAL PROGENITOR DN | 14 | 9 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-1/206 | 1484 | 1491 | 1A,m8 | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-125/351 | 1726 | 1733 | 1A,m8 | hsa-miR-125bbrain | UCCCUGAGACCCUAACUUGUGA |
| hsa-miR-125abrain | UCCCUGAGACCCUUUAACCUGUG | ||||
| miR-218 | 1262 | 1269 | 1A,m8 | hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU |
| hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU | ||||
| miR-409-3p | 1253 | 1260 | 1A,m8 | hsa-miR-409-3p | CGAAUGUUGCUCGGUGAACCCCU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.