Gene Page: HIP1
Summary ?
| GeneID | 3092 |
| Symbol | HIP1 |
| Synonyms | HIP-I|ILWEQ|SHON|SHONbeta|SHONgamma |
| Description | huntingtin interacting protein 1 |
| Reference | MIM:601767|HGNC:HGNC:4913|Ensembl:ENSG00000127946|HPRD:03461|Vega:OTTHUMG00000156050 |
| Gene type | protein-coding |
| Map location | 7q11.23 |
| Pascal p-value | 0.291 |
| Sherlock p-value | 0.832 |
| DEG p-value | DEG:Zhao_2015:p=1.55e-04:q=0.0745 |
| Fetal beta | 0.71 |
| eGene | Myers' cis & trans |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DEG:Zhao_2015 | RNA Sequencing analysis | Transcriptome sequencing and genome-wide association analyses reveal lysosomal function and actin cytoskeleton remodeling in schizophrenia and bipolar disorder. | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs869535 | chr4 | 109331957 | HIP1 | 3092 | 0.05 | trans |
Section II. Transcriptome annotation
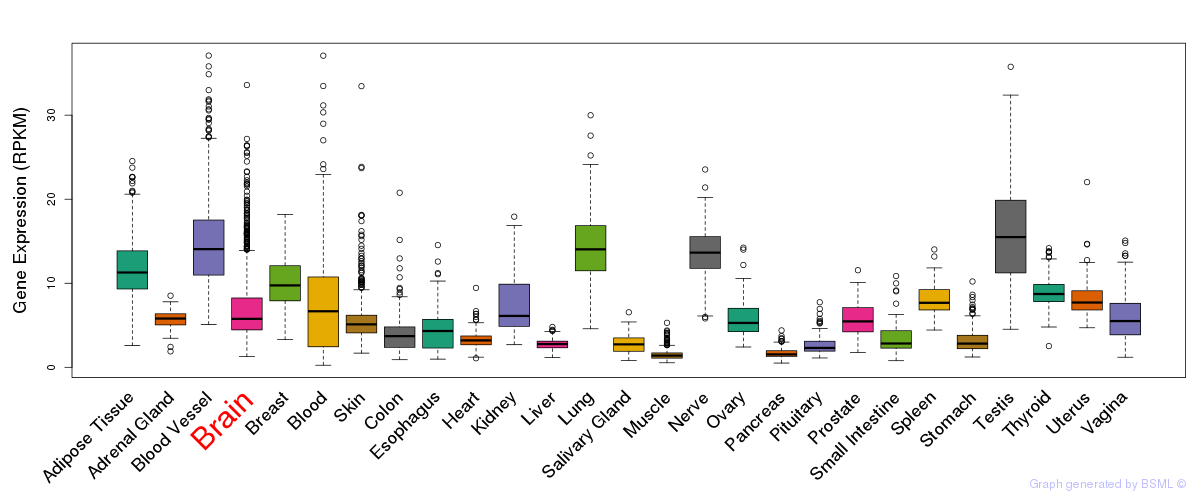
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0030276 | clathrin binding | IDA | Synap (GO term level: 4) | 11577110 |
| GO:0003779 | actin binding | IEA | - | |
| GO:0005543 | phospholipid binding | IEA | - | |
| GO:0005200 | structural constituent of cytoskeleton | TAS | 9140394 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006919 | caspase activation | IDA | 11788820 | |
| GO:0006897 | endocytosis | IDA | 11577110 |11604514 | |
| GO:0042981 | regulation of apoptosis | IDA | 11788820 | |
| GO:0048268 | clathrin coat assembly | IDA | 11577110 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0012505 | endomembrane system | IEA | - | |
| GO:0005794 | Golgi apparatus | IDA | 11788820 | |
| GO:0005856 | cytoskeleton | TAS | 9147654 | |
| GO:0005624 | membrane fraction | TAS | 9147654 | |
| GO:0005737 | cytoplasm | IDA | 11604514 | |
| GO:0016020 | membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AP2A1 | ADTAA | AP2-ALPHA | CLAPA1 | adaptor-related protein complex 2, alpha 1 subunit | - | HPRD | 11517213 |11532990|11532990 |
| AP2A1 | ADTAA | AP2-ALPHA | CLAPA1 | adaptor-related protein complex 2, alpha 1 subunit | - | HPRD,BioGRID | 11532990 |
| CASP8 | ALPS2B | CAP4 | FLICE | FLJ17672 | MACH | MCH5 | MGC78473 | caspase 8, apoptosis-related cysteine peptidase | - | HPRD,BioGRID | 11788820 |
| CLTA | LCA | clathrin, light chain (Lca) | - | HPRD | 11889126 |
| CLTB | LCB | clathrin, light chain (Lcb) | - | HPRD | 1189126 |
| CLTC | CHC | CHC17 | CLH-17 | CLTCL2 | Hc | KIAA0034 | clathrin, heavy chain (Hc) | HIP1 interacts with Clathrin-HC (Clathrin heavy chain). | BIND | 11517213 |
| CLTC | CHC | CHC17 | CLH-17 | CLTCL2 | Hc | KIAA0034 | clathrin, heavy chain (Hc) | - | HPRD,BioGRID | 11532990 |
| EIF6 | 2 | CAB | EIF3A | ITGB4BP | b | b(2)gcn | gcn | p27BBP | eukaryotic translation initiation factor 6 | Two-hybrid | BioGRID | 16169070 |
| HIP1 | ILWEQ | MGC126506 | huntingtin interacting protein 1 | - | HPRD | 11063258 |
| HIP1 | ILWEQ | MGC126506 | huntingtin interacting protein 1 | HIP1 interacts with itself. | BIND | 11063258 |
| HIP1R | FLJ14000 | FLJ27022 | HIP12 | HIP3 | ILWEQ | KIAA0655 | MGC47513 | huntingtin interacting protein 1 related | - | HPRD | 11063258 |
| HIP1R | FLJ14000 | FLJ27022 | HIP12 | HIP3 | ILWEQ | KIAA0655 | MGC47513 | huntingtin interacting protein 1 related | - | HPRD,BioGRID | 11063258 |11889126 |
| HIP1R | FLJ14000 | FLJ27022 | HIP12 | HIP3 | ILWEQ | KIAA0655 | MGC47513 | huntingtin interacting protein 1 related | HIP1 interacts with HIP12. | BIND | 11063258 |
| HTT | HD | IT15 | huntingtin | - | HPRD,BioGRID | 9147654 |11063258 |
| HTT | HD | IT15 | huntingtin | HIP1 interacts with HD. | BIND | 11063258 |
| IFT57 | ESRRBL1 | FLJ10147 | HIPPI | MHS4R2 | intraflagellar transport 57 homolog (Chlamydomonas) | - | HPRD,BioGRID | 11788820 |
| PFDN1 | PDF | PFD1 | prefoldin subunit 1 | Two-hybrid | BioGRID | 16169070 |
| RPS10 | MGC88819 | ribosomal protein S10 | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG HUNTINGTONS DISEASE | 185 | 109 | All SZGR 2.0 genes in this pathway |
| PID AR PATHWAY | 61 | 46 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 UP | 418 | 263 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION MONOCYTE DN | 54 | 38 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS UP | 238 | 144 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 DN | 378 | 231 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| WEI MIR34A TARGETS | 148 | 97 | All SZGR 2.0 genes in this pathway |
| MORI EMU MYC LYMPHOMA BY ONSET TIME DN | 17 | 9 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED UP | 183 | 111 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| DELASERNA MYOD TARGETS UP | 89 | 51 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| AMUNDSON POOR SURVIVAL AFTER GAMMA RADIATION 2G | 171 | 96 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE DN | 258 | 160 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN DN | 353 | 226 | All SZGR 2.0 genes in this pathway |
| BERNARD PPAPDC1B TARGETS DN | 58 | 39 | All SZGR 2.0 genes in this pathway |
| ZHENG GLIOBLASTOMA PLASTICITY UP | 250 | 168 | All SZGR 2.0 genes in this pathway |
| MATZUK SPERMATID DIFFERENTIATION | 37 | 26 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G2 | 27 | 17 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED | 351 | 238 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA UP | 207 | 143 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS DN | 229 | 135 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124/506 | 875 | 881 | m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-128 | 1787 | 1794 | 1A,m8 | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-27 | 1788 | 1795 | 1A,m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.