Gene Page: HNF4A
Summary ?
| GeneID | 3172 |
| Symbol | HNF4A |
| Synonyms | FRTS4|HNF4|HNF4a7|HNF4a8|HNF4a9|HNF4alpha|MODY|MODY1|NR2A1|NR2A21|TCF|TCF14 |
| Description | hepatocyte nuclear factor 4 alpha |
| Reference | MIM:600281|HGNC:HGNC:5024|Ensembl:ENSG00000101076|HPRD:02612|Vega:OTTHUMG00000032531 |
| Gene type | protein-coding |
| Map location | 20q13.12 |
| Pascal p-value | 0.919 |
| Fetal beta | -0.466 |
| eGene | Cortex Frontal Cortex BA9 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
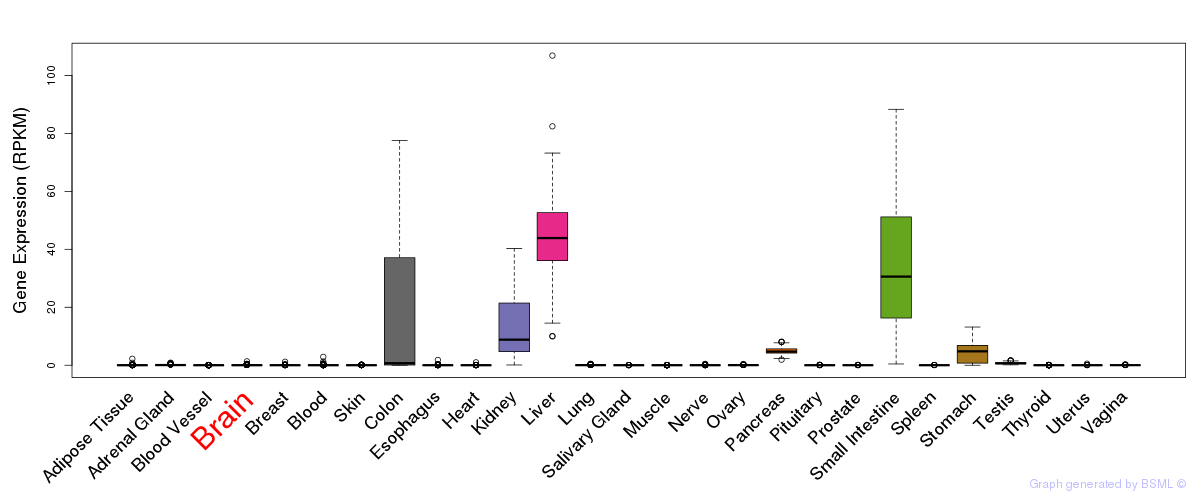
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TOP1 | 0.97 | 0.97 |
| HNRNPR | 0.97 | 0.97 |
| SFPQ | 0.97 | 0.96 |
| SYNCRIP | 0.97 | 0.97 |
| NCL | 0.96 | 0.96 |
| THRAP3 | 0.96 | 0.96 |
| HNRNPD | 0.96 | 0.96 |
| SMC3 | 0.96 | 0.96 |
| SUPT16H | 0.96 | 0.96 |
| CTCF | 0.96 | 0.96 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.75 | -0.91 |
| MT-CO2 | -0.73 | -0.90 |
| FXYD1 | -0.73 | -0.88 |
| C5orf53 | -0.73 | -0.77 |
| IFI27 | -0.73 | -0.88 |
| HLA-F | -0.73 | -0.79 |
| AF347015.27 | -0.72 | -0.86 |
| S100B | -0.72 | -0.82 |
| AF347015.33 | -0.71 | -0.86 |
| AIFM3 | -0.71 | -0.77 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005102 | receptor binding | IDA | Neurotransmitter (GO term level: 4) | 12220494 |
| GO:0003700 | transcription factor activity | IDA | 7615825 | |
| GO:0003707 | steroid hormone receptor activity | IEA | - | |
| GO:0005496 | steroid binding | IEA | - | |
| GO:0005504 | fatty acid binding | IDA | 12220494 | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0042803 | protein homodimerization activity | IDA | 12220494 | |
| GO:0046872 | metal ion binding | IEA | - | |
| GO:0043565 | sequence-specific DNA binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0010552 | positive regulation of specific transcription from RNA polymerase II promoter | IMP | 17827783 | |
| GO:0008285 | negative regulation of cell proliferation | IMP | 18163890 | |
| GO:0007596 | blood coagulation | IDA | 12911579 | |
| GO:0006805 | xenobiotic metabolic process | IMP | 17827783 | |
| GO:0006591 | ornithine metabolic process | IMP | 17827783 | |
| GO:0019216 | regulation of lipid metabolic process | IDA | 10551874 | |
| GO:0030308 | negative regulation of cell growth | IMP | 18163890 | |
| GO:0055088 | lipid homeostasis | IMP | 17827783 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005794 | Golgi apparatus | IDA | 18029348 | |
| GO:0005634 | nucleus | IDA | 7615825 | |
| GO:0005737 | cytoplasm | IDA | 14563941 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | Co-localization | BioGRID | 12944908 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | CBP interacts with the HNF-4 gene. | BIND | 15616580 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | - | HPRD,BioGRID | 9434765 |10085149 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | Affinity Capture-Western Reconstituted Complex | BioGRID | 12944908 |
| ELP3 | FLJ10422 | KAT9 | elongation protein 3 homolog (S. cerevisiae) | Elp3 interacts with the HNF-4 gene. | BIND | 15616580 |
| EXT2 | SOTV | exostoses (multiple) 2 | Affinity Capture-MS | BioGRID | 17353931 |
| FOXO1 | FKH1 | FKHR | FOXO1A | forkhead box O1 | - | HPRD,BioGRID | 12519792 |
| H3F3A | H3.3A | H3F3 | MGC87782 | MGC87783 | H3 histone, family 3A | H3F3A (Histone 3) interacts with the HNF4A gene. | BIND | 15616580 |
| HNF1A | HNF1 | LFB1 | MODY3 | TCF1 | HNF1 homeobox A | - | HPRD | 9792724 |
| MECR | CGI-63 | FASN2B | NRBF1 | mitochondrial trans-2-enoyl-CoA reductase | - | HPRD | 9795230 |
| MED1 | CRSP1 | CRSP200 | DRIP205 | DRIP230 | MGC71488 | PBP | PPARBP | PPARGBP | RB18A | TRAP220 | TRIP2 | mediator complex subunit 1 | - | HPRD,BioGRID | 12089346 |
| MED14 | CRSP150 | CRSP2 | CSRP | CXorf4 | DRIP150 | EXLM1 | MGC104513 | RGR1 | TRAP170 | mediator complex subunit 14 | - | HPRD,BioGRID | 12101254 |
| MED23 | CRSP130 | CRSP133 | CRSP3 | DKFZp434H0117 | DRIP130 | SUR2 | mediator complex subunit 23 | Affinity Capture-Western | BioGRID | 12089346 |
| NCOA1 | F-SRC-1 | KAT13A | MGC129719 | MGC129720 | NCoA-1 | RIP160 | SRC-1 | SRC1 | bHLHe42 | nuclear receptor coactivator 1 | - | HPRD,BioGRID | 9812974 |
| NCOA2 | GRIP1 | KAT13C | MGC138808 | NCoA-2 | TIF2 | nuclear receptor coactivator 2 | - | HPRD,BioGRID | 9812974 |
| NR0B2 | FLJ17090 | SHP | SHP1 | nuclear receptor subfamily 0, group B, member 2 | Reconstituted Complex Two-hybrid | BioGRID | 10594021 |
| NR2C2 | TAK1 | TR2R1 | TR4 | hTAK1 | nuclear receptor subfamily 2, group C, member 2 | - | HPRD,BioGRID | 12522137 |
| NR2F1 | COUP-TFI | EAR-3 | EAR3 | ERBAL3 | NR2F2 | SVP44 | TCFCOUP1 | TFCOUP1 | nuclear receptor subfamily 2, group F, member 1 | - | HPRD | 10652338 |
| NRBF2 | COPR1 | COPR2 | DKFZp564C1664 | FLJ30395 | NRBF-2 | nuclear receptor binding factor 2 | - | HPRD,BioGRID | 10786636 |
| PABPC4 | APP-1 | APP1 | FLJ43938 | PABP4 | iPABP | poly(A) binding protein, cytoplasmic 4 (inducible form) | Affinity Capture-MS | BioGRID | 17353931 |
| PPARGC1B | ERRL1 | PERC | PGC-1(beta) | PGC1B | peroxisome proliferator-activated receptor gamma, coactivator 1 beta | - | HPRD,BioGRID | 11733490 |
| RAD50 | RAD50-2 | hRad50 | RAD50 homolog (S. cerevisiae) | Affinity Capture-MS | BioGRID | 17353931 |
| SMAD2 | JV18 | JV18-1 | MADH2 | MADR2 | MGC22139 | MGC34440 | hMAD-2 | hSMAD2 | SMAD family member 2 | - | HPRD | 11229886 |
| SMARCA4 | BAF190 | BRG1 | FLJ39786 | SNF2 | SNF2-BETA | SNF2L4 | SNF2LB | SWI2 | hSNF2b | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 | Brg-1 interacts with the HNF-4 gene. | BIND | 15616580 |
| SNCG | BCSG1 | SR | synuclein, gamma (breast cancer-specific protein 1) | Affinity Capture-MS | BioGRID | 17353931 |
| STK16 | FLJ39635 | KRCT | MPSK | PKL12 | TSF1 | serine/threonine kinase 16 | Affinity Capture-MS | BioGRID | 17353931 |
| SUB1 | MGC102747 | P15 | PC4 | p14 | SUB1 homolog (S. cerevisiae) | - | HPRD,BioGRID | 11741883 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD,BioGRID | 11818510 |
| TRIM24 | PTC6 | RNF82 | TF1A | TIF1 | TIF1A | TIF1ALPHA | hTIF1 | tripartite motif-containing 24 | - | HPRD | 9115274 |
| ZNHIT3 | TRIP3 | zinc finger, HIT type 3 | - | HPRD,BioGRID | 11916906 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MATURITY ONSET DIABETES OF THE YOUNG | 25 | 19 | All SZGR 2.0 genes in this pathway |
| PID SMAD2 3NUCLEAR PATHWAY | 82 | 63 | All SZGR 2.0 genes in this pathway |
| PID HNF3B PATHWAY | 45 | 37 | All SZGR 2.0 genes in this pathway |
| PID HIF1 TFPATHWAY | 66 | 52 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERIC TRANSCRIPTION PATHWAY | 352 | 181 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF BETA CELL DEVELOPMENT | 30 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | 20 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | 49 | 36 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| LUCAS HNF4A TARGETS UP | 58 | 36 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO UP | 205 | 126 | All SZGR 2.0 genes in this pathway |
| MA MYELOID DIFFERENTIATION DN | 44 | 30 | All SZGR 2.0 genes in this pathway |
| YAGI AML SURVIVAL | 129 | 87 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| LABBE TARGETS OF TGFB1 AND WNT3A DN | 108 | 68 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| ZHOU PANCREATIC EXOCRINE PROGENITOR | 11 | 10 | All SZGR 2.0 genes in this pathway |
| ZHOU PANCREATIC BETA CELL | 11 | 9 | All SZGR 2.0 genes in this pathway |
| MUELLER COMMON TARGETS OF AML FUSIONS DN | 31 | 21 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS PROLIFERATION DN | 179 | 97 | All SZGR 2.0 genes in this pathway |
| YAMASHITA LIVER CANCER STEM CELL UP | 47 | 38 | All SZGR 2.0 genes in this pathway |
| YAMASHITA LIVER CANCER STEM CELL DN | 76 | 51 | All SZGR 2.0 genes in this pathway |
| WOO LIVER CANCER RECURRENCE DN | 80 | 54 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| ZEMBUTSU SENSITIVITY TO VINCRISTINE | 19 | 10 | All SZGR 2.0 genes in this pathway |
| ZEMBUTSU SENSITIVITY TO VINBLASTINE | 18 | 12 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 3 UP | 170 | 97 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 4 UP | 112 | 64 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 6 UP | 140 | 81 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 7 UP | 118 | 68 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| SERVITJA ISLET HNF1A TARGETS DN | 109 | 71 | All SZGR 2.0 genes in this pathway |