Gene Page: HP
Summary ?
| GeneID | 3240 |
| Symbol | HP |
| Synonyms | BP|HP2ALPHA2|HPA1S |
| Description | haptoglobin |
| Reference | MIM:140100|HGNC:HGNC:5141|Ensembl:ENSG00000257017|HPRD:00772|Vega:OTTHUMG00000173009 |
| Gene type | protein-coding |
| Map location | 16q22.2 |
| Pascal p-value | 0.3 |
| Sherlock p-value | 0.893 |
| Fetal beta | -0.927 |
| DMG | 1 (# studies) |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Frontal Cortex BA9 Nucleus accumbens basal ganglia Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 2 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizotypy,schizophrenias,schizotypal | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 31 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg09584711 | 16 | 72096944 | HP | 1.153E-4 | -6.894 | DMG:vanEijk_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4788579 | chr16 | 71951844 | HP | 3240 | 0 | cis | ||
| rs12708919 | chr16 | 71986994 | HP | 3240 | 0.01 | cis | ||
| rs9925462 | chr16 | 72000194 | HP | 3240 | 8.622E-9 | cis | ||
| rs1667060 | chr2 | 2945454 | HP | 3240 | 0.13 | trans | ||
| rs5009574 | chr2 | 4963368 | HP | 3240 | 0.19 | trans | ||
| rs17026940 | chr2 | 86342788 | HP | 3240 | 0.16 | trans | ||
| rs7340510 | chr2 | 134267368 | HP | 3240 | 0.01 | trans | ||
| rs6730679 | chr2 | 134282345 | HP | 3240 | 0.01 | trans | ||
| rs11929728 | chr4 | 6145405 | HP | 3240 | 0.16 | trans | ||
| rs1511040 | chr4 | 13913575 | HP | 3240 | 0.03 | trans | ||
| rs16848149 | chr4 | 73443003 | HP | 3240 | 0.01 | trans | ||
| rs1426729 | chr4 | 100671473 | HP | 3240 | 0.19 | trans | ||
| rs680474 | chr5 | 15515788 | HP | 3240 | 0.08 | trans | ||
| rs9381511 | chr6 | 12931447 | HP | 3240 | 0.02 | trans | ||
| rs12663450 | chr6 | 38135729 | HP | 3240 | 0.05 | trans | ||
| rs1381599 | chr7 | 46340766 | HP | 3240 | 0.03 | trans | ||
| rs6970701 | chr7 | 93087543 | HP | 3240 | 0.17 | trans | ||
| rs12111850 | chr7 | 93126467 | HP | 3240 | 0.17 | trans | ||
| rs6949752 | chr7 | 93138081 | HP | 3240 | 0.17 | trans | ||
| rs17165529 | chr7 | 93144582 | HP | 3240 | 0.17 | trans | ||
| rs1336572 | chr9 | 254326 | HP | 3240 | 0.03 | trans | ||
| rs13289308 | chr9 | 8971665 | HP | 3240 | 0.11 | trans | ||
| rs17038857 | chr12 | 107300920 | HP | 3240 | 4.763E-4 | trans | ||
| rs1457420 | chr14 | 66958675 | HP | 3240 | 0.17 | trans | ||
| rs17137819 | chr16 | 5372534 | HP | 3240 | 0.13 | trans | ||
| rs16958302 | chr16 | 57647432 | HP | 3240 | 0.01 | trans | ||
| rs4788579 | chr16 | 71951844 | HP | 3240 | 0.18 | trans | ||
| rs9925462 | chr16 | 72000194 | HP | 3240 | 2.078E-6 | trans | ||
| rs17001072 | chr19 | 11099774 | HP | 3240 | 0.14 | trans | ||
| rs6100270 | chr20 | 57487204 | HP | 3240 | 0.04 | trans | ||
| rs7185215 | 16 | 72034864 | HP | ENSG00000257017.4 | 4.687E-6 | 0 | -55196 | gtex_brain_ba24 |
| rs11648003 | 16 | 72052348 | HP | ENSG00000257017.4 | 9.821E-8 | 0 | -37712 | gtex_brain_ba24 |
| rs12924886 | 16 | 72075593 | HP | ENSG00000257017.4 | 4.873E-8 | 0 | -14467 | gtex_brain_ba24 |
| rs35283911 | 16 | 72078989 | HP | ENSG00000257017.4 | 4.654E-8 | 0 | -11071 | gtex_brain_ba24 |
| rs7197453 | 16 | 72079127 | HP | ENSG00000257017.4 | 2.412E-6 | 0 | -10933 | gtex_brain_ba24 |
| rs7197869 | 16 | 72079352 | HP | ENSG00000257017.4 | 2.412E-6 | 0 | -10708 | gtex_brain_ba24 |
| rs5472 | 16 | 72088467 | HP | ENSG00000257017.4 | 4.472E-7 | 0 | -1593 | gtex_brain_ba24 |
| rs8062041 | 16 | 72088964 | HP | ENSG00000257017.4 | 2.366E-6 | 0 | -1096 | gtex_brain_ba24 |
| rs3794695 | 16 | 72097827 | HP | ENSG00000257017.4 | 1.081E-7 | 0 | 7767 | gtex_brain_ba24 |
| rs34042070 | 16 | 72101525 | HP | ENSG00000257017.4 | 1.179E-7 | 0 | 11465 | gtex_brain_ba24 |
| rs2000999 | 16 | 72108093 | HP | ENSG00000257017.4 | 1.049E-7 | 0 | 18033 | gtex_brain_ba24 |
| rs2854956 | 16 | 72115238 | HP | ENSG00000257017.4 | 1.135E-6 | 0 | 25178 | gtex_brain_ba24 |
| rs9941087 | 16 | 72118324 | HP | ENSG00000257017.4 | 2.562E-6 | 0 | 28264 | gtex_brain_ba24 |
| rs397768926 | 16 | 72121552 | HP | ENSG00000257017.4 | 2.631E-6 | 0 | 31492 | gtex_brain_ba24 |
| rs12445586 | 16 | 72126276 | HP | ENSG00000257017.4 | 4.297E-6 | 0 | 36216 | gtex_brain_ba24 |
| rs1050362 | 16 | 72130815 | HP | ENSG00000257017.4 | 1.541E-6 | 0 | 40755 | gtex_brain_ba24 |
| rs34749771 | 16 | 72131740 | HP | ENSG00000257017.4 | 1.614E-6 | 0 | 41680 | gtex_brain_ba24 |
| rs6499559 | 16 | 72132064 | HP | ENSG00000257017.4 | 1.541E-6 | 0 | 42004 | gtex_brain_ba24 |
| rs2072142 | 16 | 72132713 | HP | ENSG00000257017.4 | 1.541E-6 | 0 | 42653 | gtex_brain_ba24 |
| rs2074627 | 16 | 72136769 | HP | ENSG00000257017.4 | 1.541E-6 | 0 | 46709 | gtex_brain_ba24 |
| rs12708928 | 16 | 72137227 | HP | ENSG00000257017.4 | 1.541E-6 | 0 | 47167 | gtex_brain_ba24 |
| rs2240243 | 16 | 72137561 | HP | ENSG00000257017.4 | 1.541E-6 | 0 | 47501 | gtex_brain_ba24 |
| rs12325142 | 16 | 72138112 | HP | ENSG00000257017.4 | 1.541E-6 | 0 | 48052 | gtex_brain_ba24 |
| rs2074626 | 16 | 72139184 | HP | ENSG00000257017.4 | 1.541E-6 | 0 | 49124 | gtex_brain_ba24 |
| rs2287997 | 16 | 72140553 | HP | ENSG00000257017.4 | 3.815E-8 | 0 | 50493 | gtex_brain_ba24 |
| rs9926156 | 16 | 72142895 | HP | ENSG00000257017.4 | 4.705E-6 | 0 | 52835 | gtex_brain_ba24 |
| rs2241412 | 16 | 72144284 | HP | ENSG00000257017.4 | 4.678E-6 | 0 | 54224 | gtex_brain_ba24 |
| rs12448315 | 16 | 72147242 | HP | ENSG00000257017.4 | 1.161E-6 | 0 | 57182 | gtex_brain_ba24 |
| rs6499560 | 16 | 72147666 | HP | ENSG00000257017.4 | 1.155E-6 | 0 | 57606 | gtex_brain_ba24 |
| rs8047377 | 16 | 72148339 | HP | ENSG00000257017.4 | 4.511E-6 | 0 | 58279 | gtex_brain_ba24 |
| rs12445401 | 16 | 72148419 | HP | ENSG00000257017.4 | 2.939E-8 | 0 | 58359 | gtex_brain_ba24 |
Section II. Transcriptome annotation
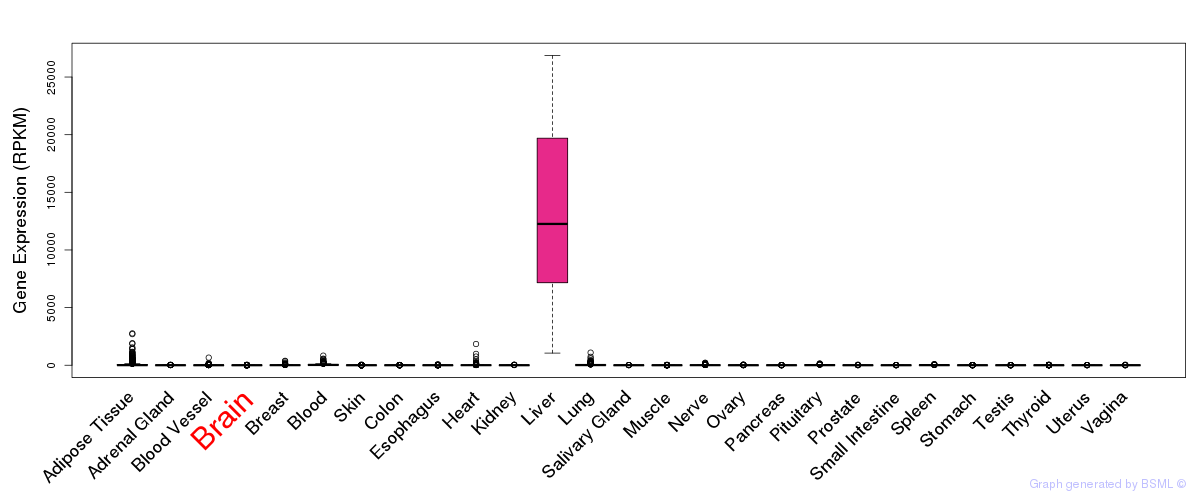
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004252 | serine-type endopeptidase activity | IEA | glutamate (GO term level: 7) | - |
| GO:0003824 | catalytic activity | IEA | - | |
| GO:0030492 | hemoglobin binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006508 | proteolysis | IEA | - | |
| GO:0006952 | defense response | TAS | 7036344 | |
| GO:0006879 | cellular iron ion homeostasis | NAS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005576 | extracellular region | NAS | 14718574 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 16D DN | 143 | 83 | All SZGR 2.0 genes in this pathway |
| LOPEZ MESOTELIOMA SURVIVAL TIME DN | 7 | 7 | All SZGR 2.0 genes in this pathway |
| BOGNI TREATMENT RELATED MYELOID LEUKEMIA UP | 30 | 19 | All SZGR 2.0 genes in this pathway |
| LOPEZ MESOTHELIOMA SURVIVAL WORST VS BEST DN | 7 | 6 | All SZGR 2.0 genes in this pathway |
| BIDUS METASTASIS DN | 161 | 93 | All SZGR 2.0 genes in this pathway |
| HAHTOLA SEZARY SYNDROM UP | 98 | 58 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF UP | 215 | 137 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 3 4WK DN | 39 | 27 | All SZGR 2.0 genes in this pathway |
| WONG ENDMETRIUM CANCER UP | 25 | 17 | All SZGR 2.0 genes in this pathway |
| HUMMERICH SKIN CANCER PROGRESSION DN | 100 | 64 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 2 | 127 | 92 | All SZGR 2.0 genes in this pathway |
| LI LUNG CANCER | 41 | 30 | All SZGR 2.0 genes in this pathway |
| HOUSTIS ROS | 36 | 29 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL AND BRAIN QTL TRANS | 185 | 114 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| HSIAO LIVER SPECIFIC GENES | 244 | 154 | All SZGR 2.0 genes in this pathway |
| NADLER OBESITY DN | 48 | 34 | All SZGR 2.0 genes in this pathway |
| GERY CEBP TARGETS | 126 | 90 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS LATE PROGENITOR | 544 | 307 | All SZGR 2.0 genes in this pathway |
| RUAN RESPONSE TO TNF UP | 12 | 10 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP | 262 | 186 | All SZGR 2.0 genes in this pathway |
| RUAN RESPONSE TO TNF TROGLITAZONE UP | 17 | 11 | All SZGR 2.0 genes in this pathway |
| ZAMORA NOS2 TARGETS DN | 96 | 71 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 6 | 189 | 112 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE UP | 156 | 92 | All SZGR 2.0 genes in this pathway |
| ZHENG IL22 SIGNALING UP | 56 | 36 | All SZGR 2.0 genes in this pathway |
| CADWELL ATG16L1 TARGETS UP | 93 | 56 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C D DN | 252 | 155 | All SZGR 2.0 genes in this pathway |
| ICHIBA GRAFT VERSUS HOST DISEASE 35D DN | 49 | 34 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA PCA1 UP | 101 | 66 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA DN | 267 | 160 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT DN | 222 | 141 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 1 | 68 | 50 | All SZGR 2.0 genes in this pathway |
| KAMIKUBO MYELOID CEBPA NETWORK | 28 | 19 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS UP | 387 | 225 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| STEGER ADIPOGENESIS UP | 21 | 16 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| YANG BCL3 TARGETS UP | 364 | 236 | All SZGR 2.0 genes in this pathway |
| VERNOCHET ADIPOGENESIS | 19 | 11 | All SZGR 2.0 genes in this pathway |
| HUANG GATA2 TARGETS UP | 149 | 96 | All SZGR 2.0 genes in this pathway |