Gene Page: PRMT2
Summary ?
| GeneID | 3275 |
| Symbol | PRMT2 |
| Synonyms | HRMT1L1 |
| Description | protein arginine methyltransferase 2 |
| Reference | MIM:601961|HGNC:HGNC:5186|Ensembl:ENSG00000160310|HPRD:09060|Vega:OTTHUMG00000048806 |
| Gene type | protein-coding |
| Map location | 21q22.3 |
| Pascal p-value | 0.102 |
| Sherlock p-value | 0.948 |
| Fetal beta | 1.009 |
| DMG | 2 (# studies) |
| eGene | Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg13271076 | 21 | 48055715 | PRMT2 | -0.019 | 0.92 | DMG:Nishioka_2013 | |
| cg04493422 | 21 | 48055366 | PRMT2 | 8.18E-9 | -0.011 | 3.87E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
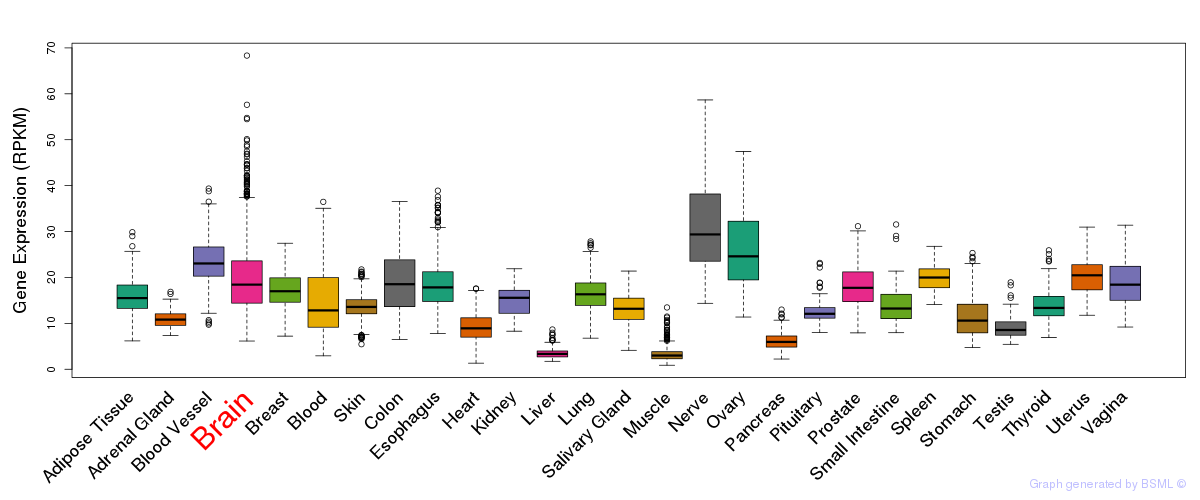
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SCP2 | 0.80 | 0.74 |
| HSP90B1 | 0.80 | 0.76 |
| NBR1 | 0.79 | 0.75 |
| HSPA9 | 0.79 | 0.72 |
| KDSR | 0.78 | 0.74 |
| GALC | 0.78 | 0.75 |
| COPB2 | 0.78 | 0.71 |
| MUT | 0.77 | 0.71 |
| C20orf43 | 0.77 | 0.69 |
| CANX | 0.77 | 0.72 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.57 | -0.36 |
| IL32 | -0.51 | -0.36 |
| IFI27 | -0.49 | -0.37 |
| AL022328.1 | -0.46 | -0.33 |
| MT-CO2 | -0.46 | -0.37 |
| C1orf61 | -0.45 | -0.29 |
| C1orf54 | -0.45 | -0.24 |
| AF347015.31 | -0.45 | -0.37 |
| CLEC2B | -0.44 | -0.21 |
| CXCL14 | -0.44 | -0.34 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BAT2 | D6S51 | D6S51E | DKFZp686D09175 | G2 | HLA-B associated transcript 2 | - | HPRD,BioGRID | 14667819 |
| DMRTB1 | - | DMRT-like family B with proline-rich C-terminal, 1 | Two-hybrid | BioGRID | 16189514 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 12039952 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | PRMT2 interacts with ER-alpha. | BIND | 12039952 |
| ESR2 | ER-BETA | ESR-BETA | ESRB | ESTRB | Erb | NR3A2 | estrogen receptor 2 (ER beta) | PRMT2 interacts with ER-beta. | BIND | 12039952 |
| FLJ12529 | FLJ39024 | MGC9315 | pre-mRNA cleavage factor I, 59 kDa subunit | Two-hybrid | BioGRID | 16189514 |
| HNRNPUL1 | E1B-AP5 | E1BAP5 | FLJ12944 | HNRPUL1 | heterogeneous nuclear ribonucleoprotein U-like 1 | - | HPRD,BioGRID | 11513728 |
| NCOA1 | F-SRC-1 | KAT13A | MGC129719 | MGC129720 | NCoA-1 | RIP160 | SRC-1 | SRC1 | bHLHe42 | nuclear receptor coactivator 1 | PRMT2 interacts with SRC-1. | BIND | 12039952 |
| NCOA6 | AIB3 | ANTP | ASC2 | HOX1.1 | HOXA7 | KIAA0181 | NRC | PRIP | RAP250 | TRBP | nuclear receptor coactivator 6 | PRMT2 interacts with PRIP. | BIND | 12039952 |
| PGR | NR3C3 | PR | progesterone receptor | PRMT2 interacts with PR. | BIND | 12039952 |
| PPARG | NR1C3 | PPARG1 | PPARG2 | PPARgamma | peroxisome proliferator-activated receptor gamma | PRMT2 interacts with PPAR-gamma. | BIND | 12039952 |
| PRMT2 | HRMT1L1 | MGC111373 | protein arginine methyltransferase 2 | PRMT2 interacts with itself. | BIND | 12039952 |
| RARA | NR1B1 | RAR | retinoic acid receptor, alpha | PRMT2 interacts with RAR. | BIND | 12039952 |
| RXRA | FLJ00280 | FLJ00318 | FLJ16020 | FLJ16733 | MGC102720 | NR2B1 | retinoid X receptor, alpha | PRMT2 interacts with RXR-alpha. | BIND | 12039952 |
| THRB | ERBA-BETA | ERBA2 | GRTH | MGC126109 | MGC126110 | NR1A2 | PRTH | THR1 | THRB1 | THRB2 | thyroid hormone receptor, beta (erythroblastic leukemia viral (v-erb-a) oncogene homolog 2, avian) | PRMT2 interacts with TR-beta. | BIND | 12039952 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DAVICIONI MOLECULAR ARMS VS ERMS UP | 332 | 228 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER DN | 406 | 230 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES CD4 DN | 116 | 71 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 UP | 256 | 159 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND DN | 225 | 163 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN | 287 | 208 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K27ME3 UP | 295 | 149 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER SURVIVAL DN | 175 | 103 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER DN | 238 | 145 | All SZGR 2.0 genes in this pathway |
| LEE EARLY T LYMPHOCYTE DN | 57 | 36 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR UP | 221 | 150 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR UP | 293 | 203 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS UP | 108 | 78 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A12 | 317 | 177 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP | 307 | 182 | All SZGR 2.0 genes in this pathway |
| CERIBELLI GENES INACTIVE AND BOUND BY NFY | 45 | 27 | All SZGR 2.0 genes in this pathway |