Gene Page: FOXN2
Summary ?
| GeneID | 3344 |
| Symbol | FOXN2 |
| Synonyms | HTLF |
| Description | forkhead box N2 |
| Reference | MIM:143089|HGNC:HGNC:5281|Ensembl:ENSG00000170802|HPRD:00882|Vega:OTTHUMG00000129168 |
| Gene type | protein-coding |
| Map location | 2p22-p16 |
| Pascal p-value | 0.002 |
| Sherlock p-value | 0.178 |
| Fetal beta | 0.369 |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cerebellar Hemisphere Cerebellum Frontal Cortex BA9 Hippocampus Nucleus accumbens basal ganglia Putamen basal ganglia Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs791272 | chr1 | 147235594 | FOXN2 | 3344 | 0.17 | trans | ||
| rs10990547 | chr9 | 105879682 | FOXN2 | 3344 | 0.16 | trans | ||
| rs113095824 | 2 | 48176656 | FOXN2 | ENSG00000170802.11 | 1.042E-6 | 0.01 | -365120 | gtex_brain_ba24 |
| rs144827198 | 2 | 48198919 | FOXN2 | ENSG00000170802.11 | 1.798E-6 | 0.01 | -342857 | gtex_brain_ba24 |
| rs115627989 | 2 | 48205690 | FOXN2 | ENSG00000170802.11 | 2.031E-6 | 0.01 | -336086 | gtex_brain_ba24 |
| rs114028693 | 2 | 48207194 | FOXN2 | ENSG00000170802.11 | 2.034E-6 | 0.01 | -334582 | gtex_brain_ba24 |
| rs80004035 | 2 | 48207554 | FOXN2 | ENSG00000170802.11 | 1.518E-6 | 0.01 | -334222 | gtex_brain_ba24 |
| rs77981979 | 2 | 48209839 | FOXN2 | ENSG00000170802.11 | 3.932E-7 | 0.01 | -331937 | gtex_brain_ba24 |
| rs77399357 | 2 | 48218576 | FOXN2 | ENSG00000170802.11 | 4.465E-7 | 0.01 | -323200 | gtex_brain_ba24 |
| rs17396122 | 2 | 48220336 | FOXN2 | ENSG00000170802.11 | 6.134E-7 | 0.01 | -321440 | gtex_brain_ba24 |
| rs17396440 | 2 | 48270256 | FOXN2 | ENSG00000170802.11 | 6.345E-7 | 0.01 | -271520 | gtex_brain_ba24 |
| rs77493474 | 2 | 48308846 | FOXN2 | ENSG00000170802.11 | 9.898E-8 | 0.01 | -232930 | gtex_brain_ba24 |
| rs77339053 | 2 | 48376101 | FOXN2 | ENSG00000170802.11 | 6.821E-8 | 0.01 | -165675 | gtex_brain_ba24 |
| rs79813800 | 2 | 48376134 | FOXN2 | ENSG00000170802.11 | 6.821E-8 | 0.01 | -165642 | gtex_brain_ba24 |
| rs78062696 | 2 | 48377720 | FOXN2 | ENSG00000170802.11 | 6.811E-8 | 0.01 | -164056 | gtex_brain_ba24 |
| rs79154515 | 2 | 48378537 | FOXN2 | ENSG00000170802.11 | 6.801E-8 | 0.01 | -163239 | gtex_brain_ba24 |
| rs75580831 | 2 | 48379382 | FOXN2 | ENSG00000170802.11 | 6.791E-8 | 0.01 | -162394 | gtex_brain_ba24 |
| rs12475492 | 2 | 48380718 | FOXN2 | ENSG00000170802.11 | 6.825E-8 | 0.01 | -161058 | gtex_brain_ba24 |
| rs76609693 | 2 | 48381731 | FOXN2 | ENSG00000170802.11 | 6.771E-8 | 0.01 | -160045 | gtex_brain_ba24 |
| rs79592437 | 2 | 48383328 | FOXN2 | ENSG00000170802.11 | 6.814E-8 | 0.01 | -158448 | gtex_brain_ba24 |
| rs75439527 | 2 | 48388666 | FOXN2 | ENSG00000170802.11 | 8.993E-8 | 0.01 | -153110 | gtex_brain_ba24 |
| rs12465922 | 2 | 48389336 | FOXN2 | ENSG00000170802.11 | 8.93E-8 | 0.01 | -152440 | gtex_brain_ba24 |
| rs62138802 | 2 | 48530981 | FOXN2 | ENSG00000170802.11 | 9.654E-8 | 0.01 | -10795 | gtex_brain_ba24 |
| rs79247094 | 2 | 48554955 | FOXN2 | ENSG00000170802.11 | 5.168E-7 | 0.01 | 13179 | gtex_brain_ba24 |
| rs79073127 | 2 | 48573895 | FOXN2 | ENSG00000170802.11 | 6.383E-7 | 0.01 | 32119 | gtex_brain_ba24 |
| rs77304590 | 2 | 48633536 | FOXN2 | ENSG00000170802.11 | 2.658E-7 | 0.01 | 91760 | gtex_brain_ba24 |
| rs116590019 | 2 | 48643248 | FOXN2 | ENSG00000170802.11 | 7.272E-7 | 0.01 | 101472 | gtex_brain_ba24 |
| rs138659652 | 2 | 48655341 | FOXN2 | ENSG00000170802.11 | 1.934E-7 | 0.01 | 113565 | gtex_brain_ba24 |
| rs72872724 | 2 | 48684796 | FOXN2 | ENSG00000170802.11 | 3.326E-7 | 0.01 | 143020 | gtex_brain_ba24 |
| rs60710459 | 2 | 48687857 | FOXN2 | ENSG00000170802.11 | 3.062E-7 | 0.01 | 146081 | gtex_brain_ba24 |
| rs139027324 | 2 | 48696459 | FOXN2 | ENSG00000170802.11 | 2.104E-7 | 0.01 | 154683 | gtex_brain_ba24 |
| rs72872744 | 2 | 48704719 | FOXN2 | ENSG00000170802.11 | 3.343E-7 | 0.01 | 162943 | gtex_brain_ba24 |
| rs57415181 | 2 | 48707841 | FOXN2 | ENSG00000170802.11 | 5.127E-7 | 0.01 | 166065 | gtex_brain_ba24 |
| rs182642283 | 2 | 48759525 | FOXN2 | ENSG00000170802.11 | 1.057E-6 | 0.01 | 217749 | gtex_brain_ba24 |
| rs143834889 | 2 | 48769903 | FOXN2 | ENSG00000170802.11 | 5.268E-7 | 0.01 | 228127 | gtex_brain_ba24 |
| rs114637089 | 2 | 48801442 | FOXN2 | ENSG00000170802.11 | 1.852E-6 | 0.01 | 259666 | gtex_brain_ba24 |
| rs77510702 | 2 | 48815306 | FOXN2 | ENSG00000170802.11 | 1.95E-6 | 0.01 | 273530 | gtex_brain_ba24 |
| rs80004035 | 2 | 48207554 | FOXN2 | ENSG00000170802.11 | 1.089E-6 | 0.04 | -334222 | gtex_brain_putamen_basal |
| rs79073127 | 2 | 48573895 | FOXN2 | ENSG00000170802.11 | 1.089E-6 | 0.04 | 32119 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
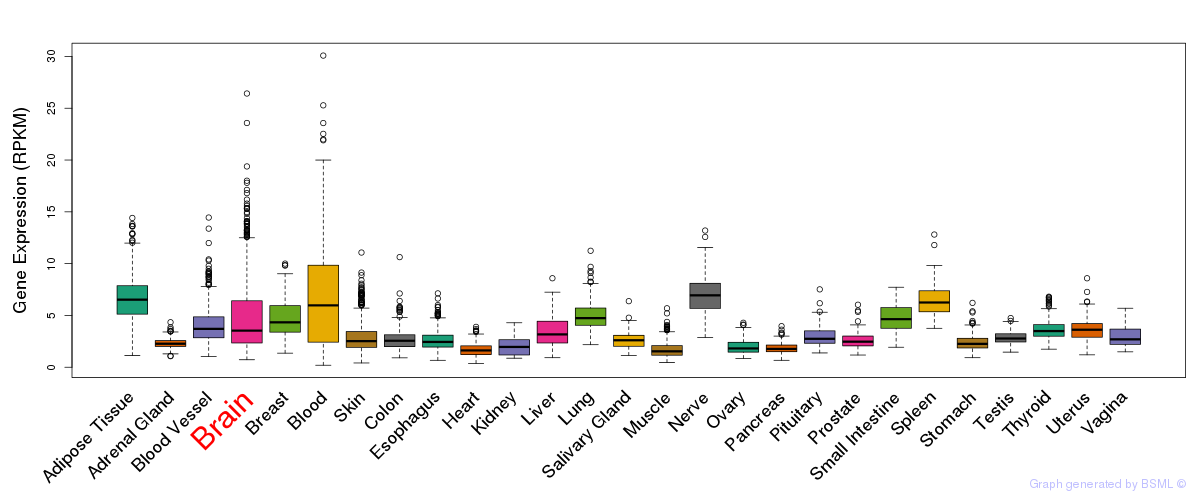
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SNAPC3 | 0.87 | 0.85 |
| CSNK2A2 | 0.85 | 0.82 |
| WDR12 | 0.85 | 0.83 |
| DNAJC18 | 0.85 | 0.85 |
| GTF3C3 | 0.85 | 0.81 |
| PAPOLA | 0.84 | 0.83 |
| BRAP | 0.84 | 0.82 |
| TIAL1 | 0.84 | 0.84 |
| SUZ12 | 0.84 | 0.81 |
| RPAP2 | 0.83 | 0.82 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.67 | -0.67 |
| AF347015.31 | -0.66 | -0.67 |
| AF347015.8 | -0.65 | -0.66 |
| IFI27 | -0.65 | -0.72 |
| AF347015.21 | -0.64 | -0.65 |
| HIGD1B | -0.64 | -0.69 |
| AF347015.2 | -0.64 | -0.65 |
| MT-CYB | -0.64 | -0.65 |
| AF347015.33 | -0.63 | -0.64 |
| FXYD1 | -0.62 | -0.69 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K9ME3 UP | 141 | 75 | All SZGR 2.0 genes in this pathway |
| HOQUE METHYLATED IN CANCER | 56 | 45 | All SZGR 2.0 genes in this pathway |
| HOFFMANN IMMATURE TO MATURE B LYMPHOCYTE DN | 50 | 36 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 UP | 397 | 206 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |