Gene Page: IGFBP7
Summary ?
| GeneID | 3490 |
| Symbol | IGFBP7 |
| Synonyms | AGM|FSTL2|IBP-7|IGFBP-7|IGFBP-7v|IGFBPRP1|MAC25|PSF|RAMSVPS|TAF |
| Description | insulin like growth factor binding protein 7 |
| Reference | MIM:602867|HGNC:HGNC:5476|Ensembl:ENSG00000163453|HPRD:04183|Vega:OTTHUMG00000128772 |
| Gene type | protein-coding |
| Map location | 4q12 |
| Pascal p-value | 0.625 |
| Sherlock p-value | 0.225 |
| Fetal beta | -0.773 |
| DMG | 1 (# studies) |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Frontal Cortex BA9 Nucleus accumbens basal ganglia Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg15924566 | 4 | 57969541 | IGFBP7 | 1.914E-4 | 0.448 | 0.035 | DMG:Wockner_2014 |
| cg01684586 | 4 | 57910078 | IGFBP7 | 3.075E-4 | 0.482 | 0.04 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2236512 | chr1 | 54519106 | IGFBP7 | 3490 | 0.17 | trans | ||
| rs12490747 | chr3 | 9092889 | IGFBP7 | 3490 | 2.638E-4 | trans | ||
| rs1709827 | chr21 | 39058204 | IGFBP7 | 3490 | 0.16 | trans | ||
| rs1714012 | 4 | 57942351 | IGFBP7 | ENSG00000163453.7 | 1.905E-6 | 0 | 34200 | gtex_brain_ba24 |
| rs1714011 | 4 | 57942630 | IGFBP7 | ENSG00000163453.7 | 1.902E-6 | 0 | 33921 | gtex_brain_ba24 |
| rs1714010 | 4 | 57942649 | IGFBP7 | ENSG00000163453.7 | 1.902E-6 | 0 | 33902 | gtex_brain_ba24 |
| rs1718847 | 4 | 57942700 | IGFBP7 | ENSG00000163453.7 | 1.902E-6 | 0 | 33851 | gtex_brain_ba24 |
| rs1718846 | 4 | 57942826 | IGFBP7 | ENSG00000163453.7 | 1.902E-6 | 0 | 33725 | gtex_brain_ba24 |
| rs9997862 | 4 | 57943318 | IGFBP7 | ENSG00000163453.7 | 1.902E-6 | 0 | 33233 | gtex_brain_ba24 |
| rs10009533 | 4 | 57943382 | IGFBP7 | ENSG00000163453.7 | 1.902E-6 | 0 | 33169 | gtex_brain_ba24 |
| rs1713981 | 4 | 57943537 | IGFBP7 | ENSG00000163453.7 | 1.956E-6 | 0 | 33014 | gtex_brain_ba24 |
| rs1713979 | 4 | 57943749 | IGFBP7 | ENSG00000163453.7 | 1.902E-6 | 0 | 32802 | gtex_brain_ba24 |
| rs1713976 | 4 | 57944503 | IGFBP7 | ENSG00000163453.7 | 9.421E-7 | 0 | 32048 | gtex_brain_ba24 |
| rs1522609 | 4 | 57944982 | IGFBP7 | ENSG00000163453.7 | 1.944E-8 | 0 | 31569 | gtex_brain_ba24 |
| rs5858411 | 4 | 57945012 | IGFBP7 | ENSG00000163453.7 | 2.673E-7 | 0 | 31539 | gtex_brain_ba24 |
| rs375959775 | 4 | 57945329 | IGFBP7 | ENSG00000163453.7 | 2.682E-7 | 0 | 31222 | gtex_brain_ba24 |
| rs1714009 | 4 | 57945371 | IGFBP7 | ENSG00000163453.7 | 1.947E-8 | 0 | 31180 | gtex_brain_ba24 |
| rs1714008 | 4 | 57945621 | IGFBP7 | ENSG00000163453.7 | 3.653E-8 | 0 | 30930 | gtex_brain_ba24 |
| rs1620509 | 4 | 57946388 | IGFBP7 | ENSG00000163453.7 | 3.662E-8 | 0 | 30163 | gtex_brain_ba24 |
| rs1714007 | 4 | 57948114 | IGFBP7 | ENSG00000163453.7 | 7.244E-7 | 0 | 28437 | gtex_brain_ba24 |
| rs1714006 | 4 | 57948145 | IGFBP7 | ENSG00000163453.7 | 7.244E-7 | 0 | 28406 | gtex_brain_ba24 |
| rs1718829 | 4 | 57948156 | IGFBP7 | ENSG00000163453.7 | 7.44E-7 | 0 | 28395 | gtex_brain_ba24 |
| rs1718858 | 4 | 57948187 | IGFBP7 | ENSG00000163453.7 | 7.244E-7 | 0 | 28364 | gtex_brain_ba24 |
| rs1718859 | 4 | 57948224 | IGFBP7 | ENSG00000163453.7 | 1.391E-7 | 0 | 28327 | gtex_brain_ba24 |
| rs1714005 | 4 | 57948823 | IGFBP7 | ENSG00000163453.7 | 1.875E-6 | 0 | 27728 | gtex_brain_ba24 |
| rs1714004 | 4 | 57949460 | IGFBP7 | ENSG00000163453.7 | 1.303E-7 | 0 | 27091 | gtex_brain_ba24 |
| rs1718861 | 4 | 57949699 | IGFBP7 | ENSG00000163453.7 | 1.475E-7 | 0 | 26852 | gtex_brain_ba24 |
| rs10015212 | 4 | 57950623 | IGFBP7 | ENSG00000163453.7 | 7.244E-7 | 0 | 25928 | gtex_brain_ba24 |
| rs12506619 | 4 | 57951010 | IGFBP7 | ENSG00000163453.7 | 7.096E-7 | 0 | 25541 | gtex_brain_ba24 |
| rs4242004 | 4 | 57953278 | IGFBP7 | ENSG00000163453.7 | 2.119E-7 | 0 | 23273 | gtex_brain_ba24 |
Section II. Transcriptome annotation
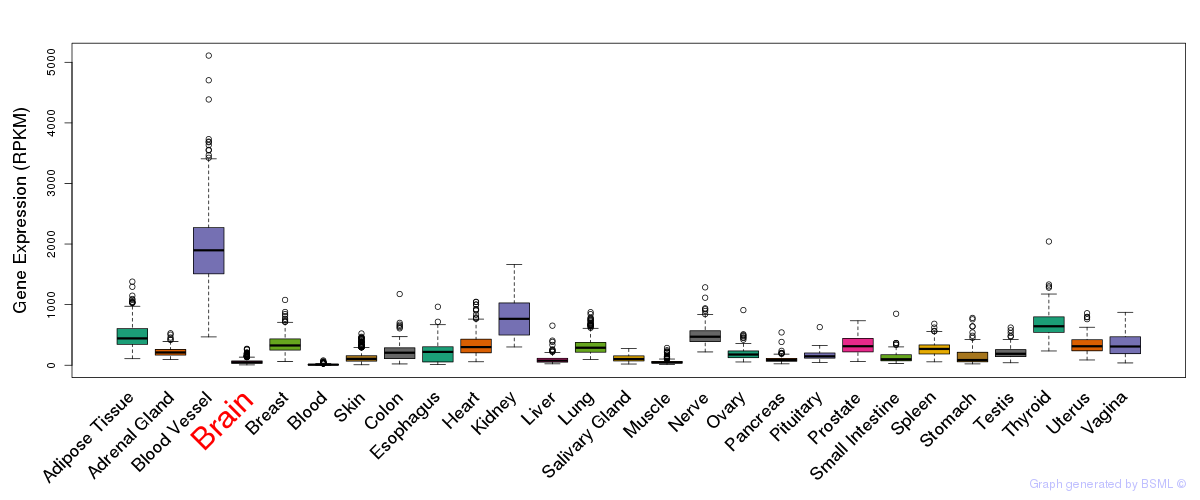
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PICCALUGA ANGIOIMMUNOBLASTIC LYMPHOMA UP | 205 | 140 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE UP | 351 | 230 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA PROGENITOR DN | 66 | 42 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA UP | 177 | 110 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN | 455 | 304 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL DN | 244 | 153 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| PEPPER CHRONIC LYMPHOCYTIC LEUKEMIA UP | 33 | 28 | All SZGR 2.0 genes in this pathway |
| AKL HTLV1 INFECTION DN | 68 | 41 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN | 362 | 238 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 UP | 380 | 236 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 UP | 418 | 263 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS DN | 232 | 139 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC2 TARGETS DN | 133 | 77 | All SZGR 2.0 genes in this pathway |
| LEE NEURAL CREST STEM CELL UP | 146 | 99 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION DN | 329 | 219 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR DN | 209 | 122 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL UP | 316 | 190 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR UP | 128 | 95 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR UP | 117 | 84 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND SERUM DEPRIVATION UP | 211 | 136 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS A UP | 191 | 128 | All SZGR 2.0 genes in this pathway |
| WANG ESOPHAGUS CANCER VS NORMAL UP | 121 | 72 | All SZGR 2.0 genes in this pathway |
| DUNNE TARGETS OF AML1 MTG8 FUSION UP | 52 | 32 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B | 392 | 251 | All SZGR 2.0 genes in this pathway |
| XU HGF SIGNALING NOT VIA AKT1 48HR DN | 20 | 12 | All SZGR 2.0 genes in this pathway |
| WEINMANN ADAPTATION TO HYPOXIA UP | 29 | 24 | All SZGR 2.0 genes in this pathway |
| WEINMANN ADAPTATION TO HYPOXIA DN | 41 | 33 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS DN | 124 | 79 | All SZGR 2.0 genes in this pathway |
| KARAKAS TGFB1 SIGNALING | 18 | 15 | All SZGR 2.0 genes in this pathway |
| INGRAM SHH TARGETS UP | 127 | 79 | All SZGR 2.0 genes in this pathway |
| MANTOVANI NFKB TARGETS UP | 43 | 33 | All SZGR 2.0 genes in this pathway |
| MANTOVANI VIRAL GPCR SIGNALING UP | 86 | 54 | All SZGR 2.0 genes in this pathway |
| ROPERO HDAC2 TARGETS | 114 | 71 | All SZGR 2.0 genes in this pathway |
| FRIDMAN SENESCENCE UP | 77 | 60 | All SZGR 2.0 genes in this pathway |
| FRIDMAN IMMORTALIZATION DN | 34 | 24 | All SZGR 2.0 genes in this pathway |
| WU CELL MIGRATION | 184 | 114 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION UP | 69 | 55 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 UP | 256 | 159 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 SIGNALING VIA CTNNB1 | 83 | 58 | All SZGR 2.0 genes in this pathway |
| CERVERA SDHB TARGETS 2 | 114 | 76 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS UP | 266 | 171 | All SZGR 2.0 genes in this pathway |
| ASTON MAJOR DEPRESSIVE DISORDER UP | 49 | 36 | All SZGR 2.0 genes in this pathway |
| SWEET KRAS TARGETS UP | 84 | 51 | All SZGR 2.0 genes in this pathway |
| LEI MYB TARGETS | 318 | 215 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 1 UP | 182 | 119 | All SZGR 2.0 genes in this pathway |
| ALCALAY AML BY NPM1 LOCALIZATION DN | 184 | 132 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED DN | 246 | 180 | All SZGR 2.0 genes in this pathway |
| BRUNO HEMATOPOIESIS | 66 | 48 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| LINDVALL IMMORTALIZED BY TERT UP | 78 | 48 | All SZGR 2.0 genes in this pathway |
| GAJATE RESPONSE TO TRABECTEDIN UP | 67 | 45 | All SZGR 2.0 genes in this pathway |
| XU GH1 EXOGENOUS TARGETS UP | 85 | 50 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV SCC UP | 123 | 75 | All SZGR 2.0 genes in this pathway |
| XU GH1 AUTOCRINE TARGETS UP | 268 | 157 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| MARCHINI TRABECTEDIN RESISTANCE DN | 49 | 34 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND ENDOTHELIUM | 66 | 47 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P2 | 79 | 55 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| CADWELL ATG16L1 TARGETS DN | 70 | 43 | All SZGR 2.0 genes in this pathway |
| HELLEBREKERS SILENCED DURING TUMOR ANGIOGENESIS | 80 | 56 | All SZGR 2.0 genes in this pathway |
| RAY TARGETS OF P210 BCR ABL FUSION DN | 16 | 11 | All SZGR 2.0 genes in this pathway |
| RUIZ TNC TARGETS UP | 153 | 107 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 DN | 315 | 201 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF DN | 235 | 144 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| ROSS LEUKEMIA WITH MLL FUSIONS | 78 | 49 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR UP | 293 | 203 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| CROONQUIST STROMAL STIMULATION UP | 60 | 42 | All SZGR 2.0 genes in this pathway |
| CROONQUIST NRAS VS STROMAL STIMULATION DN | 99 | 65 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 4 | 31 | 17 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |
| HOLLEMAN ASPARAGINASE RESISTANCE B ALL UP | 26 | 15 | All SZGR 2.0 genes in this pathway |
| NABA ECM GLYCOPROTEINS | 196 | 99 | All SZGR 2.0 genes in this pathway |
| NABA CORE MATRISOME | 275 | 148 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |