| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION
| 267 | 161 | All SZGR 2.0 genes in this pathway |
| KEGG JAK STAT SIGNALING PATHWAY
| 155 | 105 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL10 PATHWAY
| 17 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL22BP PATHWAY
| 16 | 12 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA DN
| 116 | 79 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION SUSTAINED IN GRANULOCYTE DN
| 7 | 6 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC DN
| 187 | 115 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION GRANULOCYTE DN
| 17 | 13 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL DN
| 226 | 132 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP
| 443 | 294 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS
| 1174 | 695 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B
| 392 | 251 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER CLUSTER 1
| 44 | 23 | All SZGR 2.0 genes in this pathway |
| AMUNDSON RESPONSE TO ARSENITE
| 217 | 143 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN
| 584 | 395 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK
| 1442 | 892 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3
| 1118 | 744 | All SZGR 2.0 genes in this pathway |
| MORI PRE BI LYMPHOCYTE DN
| 77 | 49 | All SZGR 2.0 genes in this pathway |
| MORI LARGE PRE BII LYMPHOCYTE DN
| 58 | 39 | All SZGR 2.0 genes in this pathway |
| MORI IMMATURE B LYMPHOCYTE UP
| 53 | 35 | All SZGR 2.0 genes in this pathway |
| MORI MATURE B LYMPHOCYTE UP
| 90 | 62 | All SZGR 2.0 genes in this pathway |
| ROSS AML WITH CBFB MYH11 FUSION
| 52 | 32 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK UP
| 87 | 58 | All SZGR 2.0 genes in this pathway |
| WIELAND UP BY HBV INFECTION
| 101 | 66 | All SZGR 2.0 genes in this pathway |
| GOLDRATH IMMUNE MEMORY
| 65 | 42 | All SZGR 2.0 genes in this pathway |
| HASLINGER B CLL WITH MUTATED VH GENES
| 18 | 14 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA CD1 AND CD2 UP
| 89 | 51 | All SZGR 2.0 genes in this pathway |
| YU MYC TARGETS DN
| 55 | 38 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP
| 390 | 242 | All SZGR 2.0 genes in this pathway |
| NIELSEN GIST AND SYNOVIAL SARCOMA DN
| 20 | 15 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP
| 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP
| 487 | 303 | All SZGR 2.0 genes in this pathway |
| MCLACHLAN DENTAL CARIES UP
| 253 | 147 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P3
| 160 | 103 | All SZGR 2.0 genes in this pathway |
| GAVIN IL2 RESPONSIVE FOXP3 TARGETS UP
| 19 | 12 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED
| 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED
| 1229 | 713 | All SZGR 2.0 genes in this pathway |
| WALLACE PROSTATE CANCER RACE UP
| 299 | 167 | All SZGR 2.0 genes in this pathway |
| MARZEC IL2 SIGNALING UP
| 115 | 80 | All SZGR 2.0 genes in this pathway |
| VILIMAS NOTCH1 TARGETS UP
| 52 | 41 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK
| 1210 | 725 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G5 DN
| 27 | 17 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS WITH HCP H3K27ME3
| 102 | 76 | All SZGR 2.0 genes in this pathway |
| KUROZUMI RESPONSE TO ONCOCYTIC VIRUS
| 44 | 30 | All SZGR 2.0 genes in this pathway |
| KUROZUMI RESPONSE TO ONCOCYTIC VIRUS AND CYCLIC RGD
| 21 | 15 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3
| 590 | 403 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP
| 857 | 456 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 UP
| 397 | 206 | All SZGR 2.0 genes in this pathway |
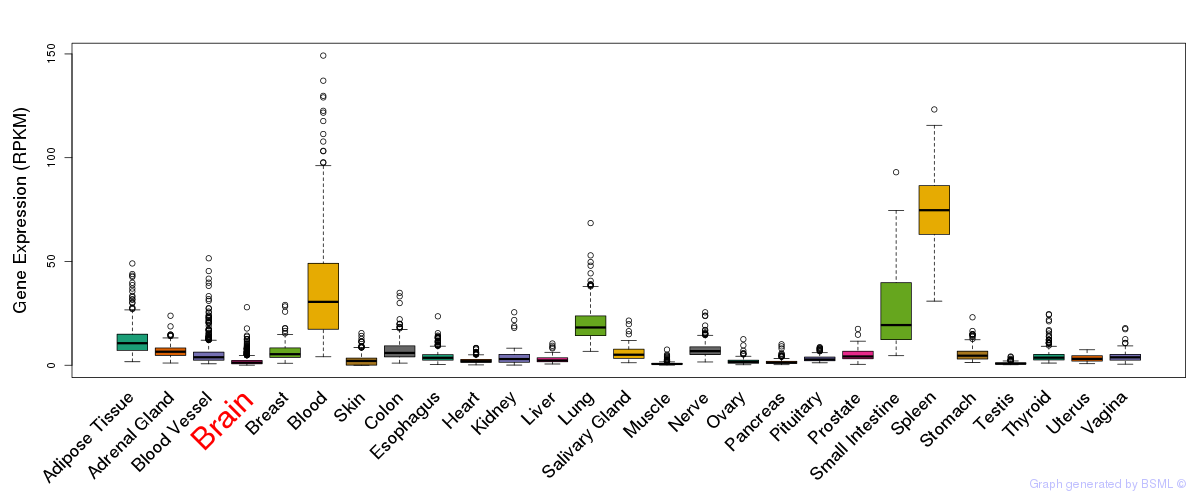
 Differentially methylated gene
Differentially methylated gene eQTL annotation
eQTL annotation