Gene Page: AQP4
Summary ?
| GeneID | 361 |
| Symbol | AQP4 |
| Synonyms | MIWC|WCH4 |
| Description | aquaporin 4 |
| Reference | MIM:600308|HGNC:HGNC:637|Ensembl:ENSG00000171885|HPRD:02631|Vega:OTTHUMG00000131955 |
| Gene type | protein-coding |
| Map location | 18q11.2-q12.1 |
| Pascal p-value | 0.127 |
| Sherlock p-value | 0.011 |
| Fetal beta | -5.934 |
| eGene | Caudate basal ganglia |
| Support | ION BALANCE CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Expression | Meta-analysis of gene expression | P value: 1.548 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
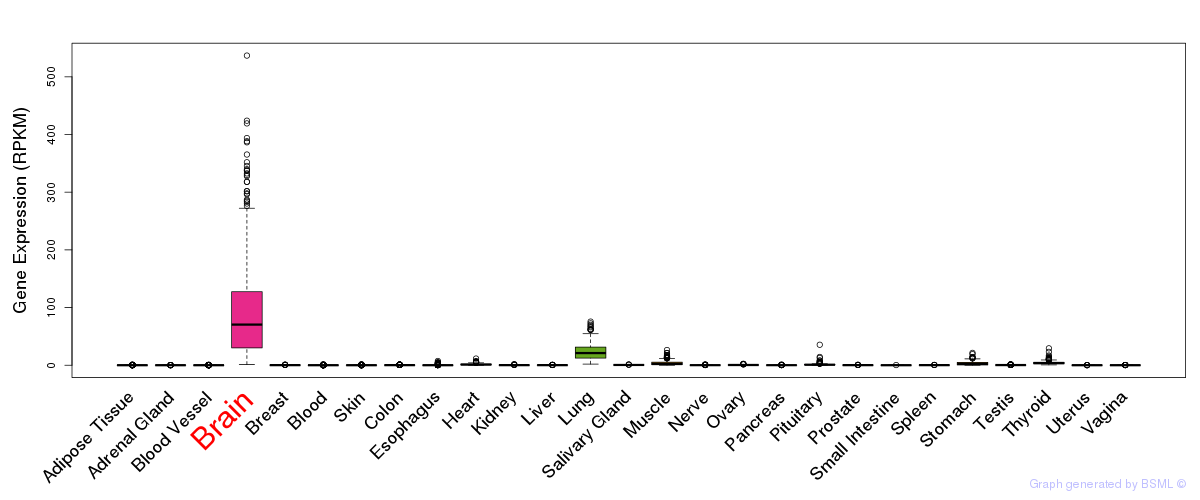
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AC134312.1 | 0.67 | 0.50 |
| MLXIPL | 0.66 | 0.58 |
| C6orf27 | 0.66 | 0.51 |
| SIRPB1 | 0.66 | 0.22 |
| HIPK4 | 0.65 | 0.57 |
| GLIS1 | 0.63 | 0.44 |
| KCNAB2 | 0.63 | 0.48 |
| SOHLH1 | 0.63 | 0.57 |
| KCNT1 | 0.63 | 0.48 |
| LYNX1 | 0.62 | 0.55 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RPS19P3 | -0.38 | -0.56 |
| RPL12 | -0.37 | -0.54 |
| C17orf45 | -0.37 | -0.54 |
| RPL14 | -0.37 | -0.52 |
| RPL18 | -0.37 | -0.56 |
| FAM36A | -0.37 | -0.36 |
| CD24 | -0.37 | -0.43 |
| RPS3P3 | -0.37 | -0.54 |
| C9orf46 | -0.37 | -0.51 |
| GTF3C6 | -0.37 | -0.40 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005372 | water transporter activity | NAS | 8855281 | |
| GO:0005372 | water transporter activity | TAS | 7559426 | |
| GO:0005215 | transporter activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007399 | nervous system development | TAS | neurite (GO term level: 5) | 7528931 |
| GO:0007588 | excretion | TAS | 7528931 | |
| GO:0006833 | water transport | NAS | 8855281 | |
| GO:0006810 | transport | IEA | - | |
| GO:0006810 | transport | TAS | 7559426 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0016020 | membrane | NAS | 8855281 | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 7528931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG VASOPRESSIN REGULATED WATER REABSORPTION | 44 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | 413 | 270 | All SZGR 2.0 genes in this pathway |
| REACTOME PASSIVE TRANSPORT BY AQUAPORINS | 11 | 7 | All SZGR 2.0 genes in this pathway |
| REACTOME AQUAPORIN MEDIATED TRANSPORT | 51 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | 44 | 30 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY DN | 367 | 220 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 DN | 242 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 DN | 162 | 116 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LPS UP | 431 | 237 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| FALVELLA SMOKERS WITH LUNG CANCER | 80 | 52 | All SZGR 2.0 genes in this pathway |
| ROPERO HDAC2 TARGETS | 114 | 71 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS UP | 126 | 84 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION NEOCORTEX UP | 83 | 66 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL LONG TERM | 302 | 191 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| MCCLUNG CREB1 TARGETS UP | 100 | 72 | All SZGR 2.0 genes in this pathway |
| LEIN ASTROCYTE MARKERS | 42 | 35 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS | 212 | 121 | All SZGR 2.0 genes in this pathway |
| KONDO PROSTATE CANCER WITH H3K27ME3 | 196 | 93 | All SZGR 2.0 genes in this pathway |
| ZHANG GATA6 TARGETS UP | 15 | 12 | All SZGR 2.0 genes in this pathway |
| LABBE TARGETS OF TGFB1 AND WNT3A UP | 111 | 70 | All SZGR 2.0 genes in this pathway |
| NUTT GBM VS AO GLIOMA UP | 46 | 34 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| HOFFMANN SMALL PRE BII TO IMMATURE B LYMPHOCYTE DN | 50 | 33 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A4 | 196 | 124 | All SZGR 2.0 genes in this pathway |
| FONTAINE THYROID TUMOR UNCERTAIN MALIGNANCY UP | 36 | 23 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 16 | 79 | 47 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 17 | 181 | 101 | All SZGR 2.0 genes in this pathway |
| BAE BRCA1 TARGETS DN | 32 | 27 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-148/152 | 11 | 17 | 1A | hsa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| miR-196 | 501 | 507 | m8 | hsa-miR-196a | UAGGUAGUUUCAUGUUGUUGG |
| hsa-miR-196b | UAGGUAGUUUCCUGUUGUUGG | ||||
| miR-29 | 1622 | 1628 | 1A | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| miR-370 | 2480 | 2486 | m8 | hsa-miR-370brain | GCCUGCUGGGGUGGAACCUGG |
| miR-384 | 1626 | 1633 | 1A,m8 | hsa-miR-384 | AUUCCUAGAAAUUGUUCAUA |
| miR-495 | 78 | 84 | 1A | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.