Gene Page: ING1
Summary ?
| GeneID | 3621 |
| Symbol | ING1 |
| Synonyms | p24ING1c|p33|p33ING1|p33ING1b|p47|p47ING1a |
| Description | inhibitor of growth family member 1 |
| Reference | MIM:601566|HGNC:HGNC:6062|Ensembl:ENSG00000153487|HPRD:03337|Vega:OTTHUMG00000017346 |
| Gene type | protein-coding |
| Map location | 13q34 |
| Pascal p-value | 0.05 |
| Sherlock p-value | 0.109 |
| Fetal beta | 1.13 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| CV:PheWAS | Phenome-wide association studies (PheWAS) | 157 SNPs associated with schizophrenia | 0 |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 CV:PheWAS
CV:PheWAS
| SNP ID | Chromosome | Position | Allele | P | Function | Gene | Up/Down Distance |
|---|
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg02550602 | 13 | 111366822 | ING1 | 7.62E-8 | 0.005 | 1.8E-5 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
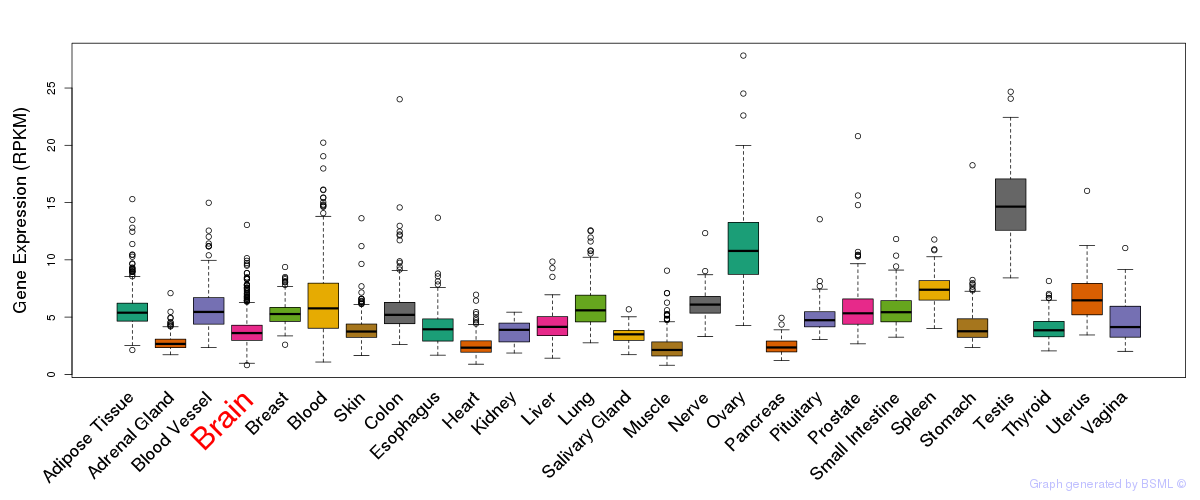
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AC068888.1 | 0.94 | 0.88 |
| POLR1D | 0.94 | 0.92 |
| DPH5 | 0.93 | 0.93 |
| TINP1 | 0.92 | 0.88 |
| IGBP1 | 0.92 | 0.91 |
| NPM1 | 0.92 | 0.88 |
| CCT2 | 0.91 | 0.91 |
| EIF2A | 0.91 | 0.90 |
| RSL1D1 | 0.91 | 0.89 |
| TEX10 | 0.91 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SLC9A3R2 | -0.61 | -0.69 |
| HLA-F | -0.59 | -0.79 |
| AIFM3 | -0.58 | -0.78 |
| AL132868.3 | -0.57 | -0.75 |
| ZBTB47 | -0.57 | -0.62 |
| APOL1 | -0.56 | -0.79 |
| FBXO2 | -0.56 | -0.70 |
| ATP10A | -0.56 | -0.74 |
| LHPP | -0.55 | -0.61 |
| APOL3 | -0.55 | -0.78 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTL6A | ACTL6 | Arp4 | BAF53A | INO80K | MGC5382 | actin-like 6A | Affinity Capture-MS | BioGRID | 11784859 |
| ARID1A | B120 | BAF250 | BAF250a | BM029 | C1orf4 | P270 | SMARCF1 | AT rich interactive domain 1A (SWI-like) | Affinity Capture-MS | BioGRID | 11784859 |
| C20orf20 | Eaf7 | FLJ10914 | MRG15BP | MRGBP | URCC4 | chromosome 20 open reading frame 20 | - | HPRD | 12963728 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | Affinity Capture-Western | BioGRID | 12015309 |
| DMAP1 | DKFZp686L09142 | DNMAP1 | DNMTAP1 | EAF2 | FLJ11543 | KIAA1425 | SWC4 | DNA methyltransferase 1 associated protein 1 | - | HPRD,BioGRID | 14665632 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | - | HPRD,BioGRID | 12015309 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 11784859 |12015309 |
| HDAC2 | RPD3 | YAF1 | histone deacetylase 2 | Affinity Capture-MS | BioGRID | 11784859 |
| HIST2H3A | H3/n | H3/o | histone cluster 2, H3a | Biochemical Activity | BioGRID | 12015309 |
| HIST4H4 | H4/p | MGC24116 | histone cluster 4, H4 | Biochemical Activity | BioGRID | 12015309 |
| KAT2B | CAF | P | P/CAF | PCAF | K(lysine) acetyltransferase 2B | - | HPRD,BioGRID | 12015309 |
| PCNA | MGC8367 | proliferating cell nuclear antigen | - | HPRD,BioGRID | 11682605 |12015309 |
| RBBP4 | NURF55 | RBAP48 | retinoblastoma binding protein 4 | Affinity Capture-MS | BioGRID | 11784859 |
| RBBP7 | MGC138867 | MGC138868 | RbAp46 | retinoblastoma binding protein 7 | Affinity Capture-MS | BioGRID | 11784859 |
| SAP30 | - | Sin3A-associated protein, 30kDa | - | HPRD,BioGRID | 11784859 |
| SIN3A | DKFZp434K2235 | FLJ90319 | KIAA0700 | SIN3 homolog A, transcription regulator (yeast) | Affinity Capture-Western | BioGRID | 11784859 |
| SMARCA4 | BAF190 | BRG1 | FLJ39786 | SNF2 | SNF2-BETA | SNF2L4 | SNF2LB | SWI2 | hSNF2b | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 11784859 |
| SMARCB1 | BAF47 | INI1 | RDT | SNF5 | SNF5L1 | Sfh1p | Snr1 | hSNFS | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1 | Affinity Capture-MS | BioGRID | 11784859 |
| SMARCC1 | BAF155 | CRACC1 | Rsc8 | SRG3 | SWI3 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 11784859 |
| SMARCC2 | BAF170 | CRACC2 | Rsc8 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 | Affinity Capture-MS | BioGRID | 11784859 |
| SMARCD1 | BAF60A | CRACD1 | Rsc6p | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 | Affinity Capture-MS | BioGRID | 11784859 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD,BioGRID | 9440695 |12208736 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | p53 interacts with ING1b. This interaction was modelled on a demonstrated interaction between p53 from an unspecified species and human ING1b. | BIND | 12208736 |
| TRRAP | FLJ10671 | PAF350/400 | PAF400 | STAF40 | TR-AP | Tra1 | transformation/transcription domain-associated protein | Affinity Capture-Western | BioGRID | 12015309 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 UP | 329 | 196 | All SZGR 2.0 genes in this pathway |
| ROVERSI GLIOMA COPY NUMBER DN | 54 | 37 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| SCHWAB TARGETS OF BMYB POLYMORPHIC VARIANTS UP | 13 | 9 | All SZGR 2.0 genes in this pathway |
| FRIDMAN SENESCENCE UP | 77 | 60 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER EXPRESSION BY COPY NUMBER | 100 | 62 | All SZGR 2.0 genes in this pathway |
| WILLERT WNT SIGNALING | 24 | 13 | All SZGR 2.0 genes in this pathway |
| PEART HDAC PROLIFERATION CLUSTER UP | 57 | 35 | All SZGR 2.0 genes in this pathway |
| SASAKI ADULT T CELL LEUKEMIA | 176 | 122 | All SZGR 2.0 genes in this pathway |
| FAELT B CLL WITH VH REARRANGEMENTS UP | 48 | 34 | All SZGR 2.0 genes in this pathway |
| HADDAD B LYMPHOCYTE PROGENITOR | 293 | 193 | All SZGR 2.0 genes in this pathway |
| FAELT B CLL WITH VH3 21 DN | 49 | 29 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR UP | 156 | 101 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 10HR UP | 101 | 69 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY DEACETYLATION IN PANCREATIC CANCER | 50 | 30 | All SZGR 2.0 genes in this pathway |
| STEARMAN LUNG CANCER EARLY VS LATE UP | 125 | 89 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS UP | 317 | 208 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR BOUND ES | 462 | 273 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE VIA P38 COMPLETE | 227 | 151 | All SZGR 2.0 genes in this pathway |