Gene Page: ITGAX
Summary ?
| GeneID | 3687 |
| Symbol | ITGAX |
| Synonyms | CD11C|SLEB6 |
| Description | integrin subunit alpha X |
| Reference | MIM:151510|HGNC:HGNC:6152|Ensembl:ENSG00000140678|HPRD:01051|Vega:OTTHUMG00000132465 |
| Gene type | protein-coding |
| Map location | 16p11.2 |
| Pascal p-value | 0.196 |
| Sherlock p-value | 0.029 |
| Fetal beta | -1.015 |
| eGene | Myers' cis & trans |
| Support | G2Cdb.humanPSD G2Cdb.humanPSP |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01775 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10031882 | chr4 | 160747054 | ITGAX | 3687 | 0.09 | trans | ||
| rs7830259 | chr8 | 14087643 | ITGAX | 3687 | 0.15 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
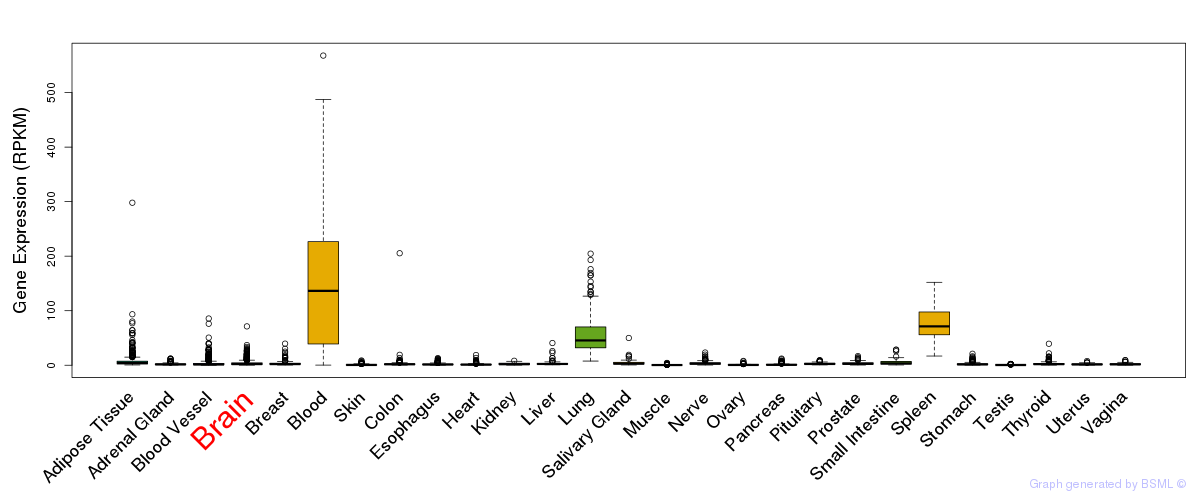
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PYCR2 | 0.78 | 0.73 |
| SIDT2 | 0.78 | 0.71 |
| PIGV | 0.77 | 0.73 |
| TMEM129 | 0.77 | 0.75 |
| TMEM214 | 0.76 | 0.68 |
| SLC6A8 | 0.75 | 0.71 |
| FICD | 0.74 | 0.71 |
| SLC41A1 | 0.74 | 0.74 |
| SLC44A2 | 0.74 | 0.75 |
| RECQL5 | 0.73 | 0.68 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FAM159B | -0.45 | -0.48 |
| AF347015.21 | -0.41 | -0.19 |
| AC087071.1 | -0.40 | -0.28 |
| AL050337.1 | -0.38 | -0.25 |
| RP9P | -0.38 | -0.36 |
| SPDYA | -0.35 | -0.25 |
| NOX1 | -0.34 | -0.25 |
| RPL13AP22 | -0.34 | -0.43 |
| SYCP3 | -0.34 | -0.22 |
| DDTL | -0.34 | -0.24 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000287 | magnesium ion binding | IEA | - | |
| GO:0004872 | receptor activity | IEA | - | |
| GO:0004872 | receptor activity | TAS | 2303426 | |
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007155 | cell adhesion | IEA | - | |
| GO:0007155 | cell adhesion | TAS | 2303426 | |
| GO:0007229 | integrin-mediated signaling pathway | IEA | - | |
| GO:0009887 | organ morphogenesis | TAS | 3327687 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0009897 | external side of plasma membrane | IEA | - | |
| GO:0008305 | integrin complex | TAS | 2303426 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| BIOCARTA DC PATHWAY | 22 | 20 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN CS PATHWAY | 26 | 16 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN2 PATHWAY | 29 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | 91 | 65 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRIN CELL SURFACE INTERACTIONS | 79 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 1 UP | 276 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 UP | 380 | 236 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 UP | 418 | 263 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 UP | 341 | 197 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF UP | 215 | 137 | All SZGR 2.0 genes in this pathway |
| APPIERTO RESPONSE TO FENRETINIDE UP | 38 | 26 | All SZGR 2.0 genes in this pathway |
| AMUNDSON RESPONSE TO ARSENITE | 217 | 143 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| GOLUB ALL VS AML DN | 24 | 14 | All SZGR 2.0 genes in this pathway |
| ROSS AML WITH MLL FUSIONS | 78 | 45 | All SZGR 2.0 genes in this pathway |
| BROWN MYELOID CELL DEVELOPMENT UP | 165 | 100 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS DN | 366 | 238 | All SZGR 2.0 genes in this pathway |
| YAGI AML FAB MARKERS | 191 | 131 | All SZGR 2.0 genes in this pathway |
| STEARMAN TUMOR FIELD EFFECT UP | 36 | 22 | All SZGR 2.0 genes in this pathway |
| STEARMAN LUNG CANCER EARLY VS LATE DN | 61 | 39 | All SZGR 2.0 genes in this pathway |
| PODAR RESPONSE TO ADAPHOSTIN UP | 147 | 98 | All SZGR 2.0 genes in this pathway |
| GOLDRATH ANTIGEN RESPONSE | 346 | 192 | All SZGR 2.0 genes in this pathway |
| LINDSTEDT DENDRITIC CELL MATURATION D | 68 | 44 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER UP | 288 | 168 | All SZGR 2.0 genes in this pathway |