Gene Page: ITGB2
Summary ?
| GeneID | 3689 |
| Symbol | ITGB2 |
| Synonyms | CD18|LAD|LCAMB|LFA-1|MAC-1|MF17|MFI7 |
| Description | integrin subunit beta 2 |
| Reference | MIM:600065|HGNC:HGNC:6155|Ensembl:ENSG00000160255|HPRD:02506|Vega:OTTHUMG00000090257 |
| Gene type | protein-coding |
| Map location | 21q22.3 |
| Pascal p-value | 0.656 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 1 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 5.707 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg04790129 | 21 | 46340810 | ITGB2 | 9.067E-4 | -4.48 | DMG:vanEijk_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2892394 | chr10 | 1740277 | ITGB2 | 3689 | 0.18 | trans |
Section II. Transcriptome annotation
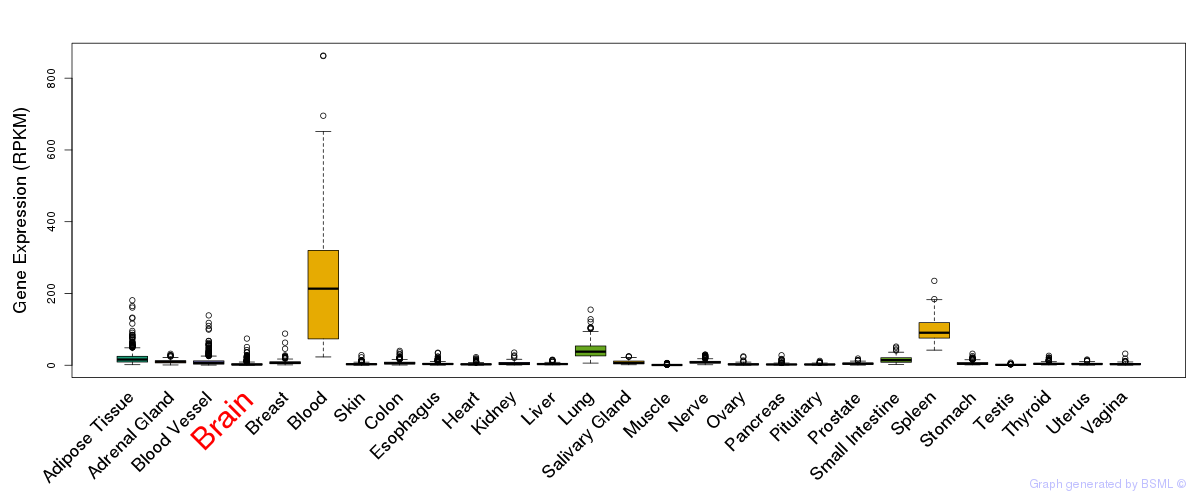
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SPIRE2 | 0.90 | 0.90 |
| C7orf51 | 0.89 | 0.90 |
| MAST1 | 0.89 | 0.90 |
| KIFC3 | 0.89 | 0.90 |
| ZNF512B | 0.88 | 0.92 |
| ABCC10 | 0.88 | 0.88 |
| CIC | 0.88 | 0.90 |
| AC010536.2 | 0.87 | 0.89 |
| ANO8 | 0.87 | 0.87 |
| SNAPC4 | 0.87 | 0.90 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.67 | -0.73 |
| S100B | -0.65 | -0.72 |
| COPZ2 | -0.65 | -0.71 |
| AC021016.1 | -0.64 | -0.76 |
| AF347015.27 | -0.64 | -0.71 |
| B2M | -0.64 | -0.70 |
| MT-CO2 | -0.64 | -0.72 |
| PSMB9 | -0.64 | -0.69 |
| AF347015.33 | -0.63 | -0.68 |
| C5orf53 | -0.63 | -0.64 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004872 | receptor activity | IEA | - | |
| GO:0005488 | binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 12885943 | |
| GO:0019901 | protein kinase binding | IPI | 12885943 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007160 | cell-matrix adhesion | IEA | - | |
| GO:0007155 | cell adhesion | IEA | - | |
| GO:0007155 | cell adhesion | TAS | 1694220 | |
| GO:0007267 | cell-cell signaling | NAS | 12885943 | |
| GO:0007229 | integrin-mediated signaling pathway | IEA | - | |
| GO:0007229 | integrin-mediated signaling pathway | NAS | 12885943 | |
| GO:0008360 | regulation of cell shape | NAS | 12885943 | |
| GO:0006954 | inflammatory response | NAS | 12885943 | |
| GO:0006915 | apoptosis | NAS | 12885943 | |
| GO:0007159 | leukocyte adhesion | IDA | 12885943 | |
| GO:0030593 | neutrophil chemotaxis | IDA | 12885943 | |
| GO:0050730 | regulation of peptidyl-tyrosine phosphorylation | IDA | 12885943 | |
| GO:0045123 | cellular extravasation | IEA | - | |
| GO:0050798 | activated T cell proliferation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005624 | membrane fraction | IEA | - | |
| GO:0005737 | cytoplasm | IDA | 18029348 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | EXP | 11857637 | |
| GO:0005886 | plasma membrane | IDA | 18029348 | |
| GO:0008305 | integrin complex | IEA | - | |
| GO:0008305 | integrin complex | NAS | 12885943 | |
| GO:0045121 | membrane raft | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CD14 | - | CD14 molecule | - | HPRD | 11745332 |
| CD226 | DNAM-1 | DNAM1 | PTA1 | TLiSA1 | CD226 molecule | - | HPRD,BioGRID | 10591186 |
| CD82 | 4F9 | C33 | GR15 | IA4 | KAI1 | R2 | SAR2 | ST6 | TSPAN27 | CD82 molecule | - | HPRD | 10602019 |
| CYTH1 | B2-1 | CYTOHESIN-1 | D17S811E | FLJ34050 | FLJ41900 | PSCD1 | SEC7 | cytohesin 1 | - | HPRD,BioGRID | 10835351 |
| CYTH1 | B2-1 | CYTOHESIN-1 | D17S811E | FLJ34050 | FLJ41900 | PSCD1 | SEC7 | cytohesin 1 | Integrin beta-2 interacts with cytohesin-1. | BIND | 9765275 |
| ESM1 | endocan | endothelial cell-specific molecule 1 | - | HPRD | 11544294 |
| FHL2 | AAG11 | DRAL | SLIM3 | four and a half LIM domains 2 | - | HPRD,BioGRID | 10906324 |
| FUT4 | CD15 | ELFT | FCT3A | FUC-TIV | fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) | - | HPRD | 3305706 |
| GNB2L1 | Gnb2-rs1 | H12.3 | HLC-7 | PIG21 | RACK1 | guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 | - | HPRD,BioGRID | 9442085 |
| HP | BP | HP2-ALPHA-2 | HPA1S | MGC111141 | haptoglobin | - | HPRD | 11967116 |
| ICAM1 | BB2 | CD54 | P3.58 | intercellular adhesion molecule 1 | - | HPRD | 7642561 |11279101 |
| ICAM1 | BB2 | CD54 | P3.58 | intercellular adhesion molecule 1 | in vitro in vivo Reconstituted Complex | BioGRID | 7642561 |10352278 |11279101 |
| ICAM2 | CD102 | intercellular adhesion molecule 2 | - | HPRD | 9786897 |
| ICAM3 | CD50 | CDW50 | ICAM-R | intercellular adhesion molecule 3 | - | HPRD,BioGRID | 7822326 |11353828 |
| ICAM4 | CD242 | LW | intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) | - | HPRD | 10846180 |
| ICAM5 | TLCN | TLN | intercellular adhesion molecule 5, telencephalin | - | HPRD,BioGRID | 8995416 |
| ILK | DKFZp686F1765 | P59 | integrin-linked kinase | - | HPRD | 9736715 |
| ITGAD | ADB2 | CD11D | FLJ39841 | integrin, alpha D | - | HPRD,BioGRID | 8777714 |
| ITGAL | CD11A | LFA-1 | LFA1A | integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) | Affinity Capture-Western | BioGRID | 9442085 |
| ITGAM | CD11B | CR3A | MAC-1 | MAC1A | MGC117044 | MO1A | SLEB6 | integrin, alpha M (complement component 3 receptor 3 subunit) | - | HPRD,BioGRID | 2454931 |
| ITGAX | CD11C | SLEB6 | integrin, alpha X (complement component 3 receptor 4 subunit) | - | HPRD,BioGRID | 11937543 |
| KNG1 | BDK | KNG | kininogen 1 | - | HPRD,BioGRID | 10845911 |
| PLAUR | CD87 | UPAR | URKR | plasminogen activator, urokinase receptor | Affinity Capture-Western | BioGRID | 10388537 |
| PRTN3 | ACPA | AGP7 | C-ANCA | MBT | P29 | PR-3 | proteinase 3 | - | HPRD | 12960243 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | - | HPRD | 8649427 |
| PTK2B | CADTK | CAKB | FADK2 | FAK2 | FRNK | PKB | PTK | PYK2 | RAFTK | PTK2B protein tyrosine kinase 2 beta | - | HPRD,BioGRID | 10961871 |
| RANBP9 | RANBPM | RAN binding protein 9 | RanBPM interacts with beta-2 integrin LFA-1. | BIND | 14722085 |
| SYK | DKFZp313N1010 | FLJ25043 | FLJ37489 | spleen tyrosine kinase | Affinity Capture-Western | BioGRID | 10961871 |
| TLN1 | ILWEQ | KIAA1027 | TLN | talin 1 | - | HPRD | 2150484 |
| VNN2 | FOAP-4 | GPI-80 | vanin 2 | - | HPRD,BioGRID | 12056825 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL ADHESION MOLECULES CAMS | 134 | 93 | All SZGR 2.0 genes in this pathway |
| KEGG NATURAL KILLER CELL MEDIATED CYTOTOXICITY | 137 | 92 | All SZGR 2.0 genes in this pathway |
| KEGG LEUKOCYTE TRANSENDOTHELIAL MIGRATION | 118 | 78 | All SZGR 2.0 genes in this pathway |
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| KEGG LEISHMANIA INFECTION | 72 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG VIRAL MYOCARDITIS | 73 | 58 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GRANULOCYTES PATHWAY | 14 | 9 | All SZGR 2.0 genes in this pathway |
| BIOCARTA LYM PATHWAY | 11 | 6 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BLYMPHOCYTE PATHWAY | 11 | 8 | All SZGR 2.0 genes in this pathway |
| BIOCARTA LAIR PATHWAY | 17 | 12 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CTL PATHWAY | 15 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MONOCYTE PATHWAY | 11 | 5 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TCYTOTOXIC PATHWAY | 14 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA THELPER PATHWAY | 14 | 11 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN CS PATHWAY | 26 | 16 | All SZGR 2.0 genes in this pathway |
| PID AMB2 NEUTROPHILS PATHWAY | 41 | 32 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN2 PATHWAY | 29 | 18 | All SZGR 2.0 genes in this pathway |
| PID UPA UPAR PATHWAY | 42 | 30 | All SZGR 2.0 genes in this pathway |
| PID CXCR3 PATHWAY | 43 | 34 | All SZGR 2.0 genes in this pathway |
| PID HIF1 TFPATHWAY | 66 | 52 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | 70 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | 91 | 65 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRIN CELL SURFACE INTERACTIONS | 79 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 UP | 380 | 236 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 UP | 418 | 263 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL DN | 226 | 132 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF UP | 215 | 137 | All SZGR 2.0 genes in this pathway |
| HERNANDEZ MITOTIC ARREST BY DOCETAXEL 2 DN | 19 | 13 | All SZGR 2.0 genes in this pathway |
| SHI SPARC TARGETS DN | 13 | 8 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO BEXAROTENE DN | 29 | 19 | All SZGR 2.0 genes in this pathway |
| FERRANDO TAL1 NEIGHBORS | 21 | 15 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK UP | 87 | 58 | All SZGR 2.0 genes in this pathway |
| BROWN MYELOID CELL DEVELOPMENT UP | 165 | 100 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS DN | 366 | 238 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND DN | 225 | 163 | All SZGR 2.0 genes in this pathway |
| LIAN NEUTROPHIL GRANULE CONSTITUENTS | 25 | 13 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA DN | 41 | 27 | All SZGR 2.0 genes in this pathway |
| ZHOU TNF SIGNALING 4HR | 54 | 36 | All SZGR 2.0 genes in this pathway |
| RAMALHO STEMNESS DN | 74 | 55 | All SZGR 2.0 genes in this pathway |
| NIELSEN GIST AND SYNOVIAL SARCOMA DN | 20 | 15 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA RESPONSE TO TGFB1 C2 | 25 | 18 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA EARLY RESPONSE TO TGFB1 | 58 | 43 | All SZGR 2.0 genes in this pathway |
| MCLACHLAN DENTAL CARIES DN | 245 | 144 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| MCLACHLAN DENTAL CARIES UP | 253 | 147 | All SZGR 2.0 genes in this pathway |
| WALLACE PROSTATE CANCER RACE UP | 299 | 167 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA UP | 259 | 185 | All SZGR 2.0 genes in this pathway |
| WU SILENCED BY METHYLATION IN BLADDER CANCER | 55 | 42 | All SZGR 2.0 genes in this pathway |
| ICHIBA GRAFT VERSUS HOST DISEASE 35D UP | 131 | 79 | All SZGR 2.0 genes in this pathway |
| LINDSTEDT DENDRITIC CELL MATURATION D | 68 | 44 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 DN | 240 | 153 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| SWEET KRAS ONCOGENIC SIGNATURE | 89 | 56 | All SZGR 2.0 genes in this pathway |
| LEE DIFFERENTIATING T LYMPHOCYTE | 200 | 115 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| LIU VAV3 PROSTATE CARCINOGENESIS UP | 89 | 61 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER UP | 288 | 168 | All SZGR 2.0 genes in this pathway |
| YAMANAKA GLIOBLASTOMA SURVIVAL DN | 9 | 7 | All SZGR 2.0 genes in this pathway |
| OKAMOTO LIVER CANCER MULTICENTRIC OCCURRENCE UP | 25 | 15 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S1 | 237 | 159 | All SZGR 2.0 genes in this pathway |
| NIELSEN SYNOVIAL SARCOMA DN | 20 | 13 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 30MIN DN | 150 | 99 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA MESENCHYMAL | 216 | 130 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 2 DN | 9 | 8 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 5 DN | 50 | 31 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS UP | 259 | 159 | All SZGR 2.0 genes in this pathway |
| SERVITJA ISLET HNF1A TARGETS UP | 163 | 111 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 6 | 29 | 20 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |