Gene Page: ITPK1
Summary ?
| GeneID | 3705 |
| Symbol | ITPK1 |
| Synonyms | ITRPK1 |
| Description | inositol-tetrakisphosphate 1-kinase |
| Reference | MIM:601838|HGNC:HGNC:6177|Ensembl:ENSG00000100605|HPRD:03501|Vega:OTTHUMG00000171226 |
| Gene type | protein-coding |
| Map location | 14q31 |
| Pascal p-value | 0.672 |
| Sherlock p-value | 0.997 |
| Fetal beta | -1.732 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg10527288 | 14 | 93582455 | ITPK1 | 3.362E-4 | -0.348 | 0.041 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs9567401 | chr13 | 44860076 | ITPK1 | 3705 | 0.18 | trans |
Section II. Transcriptome annotation
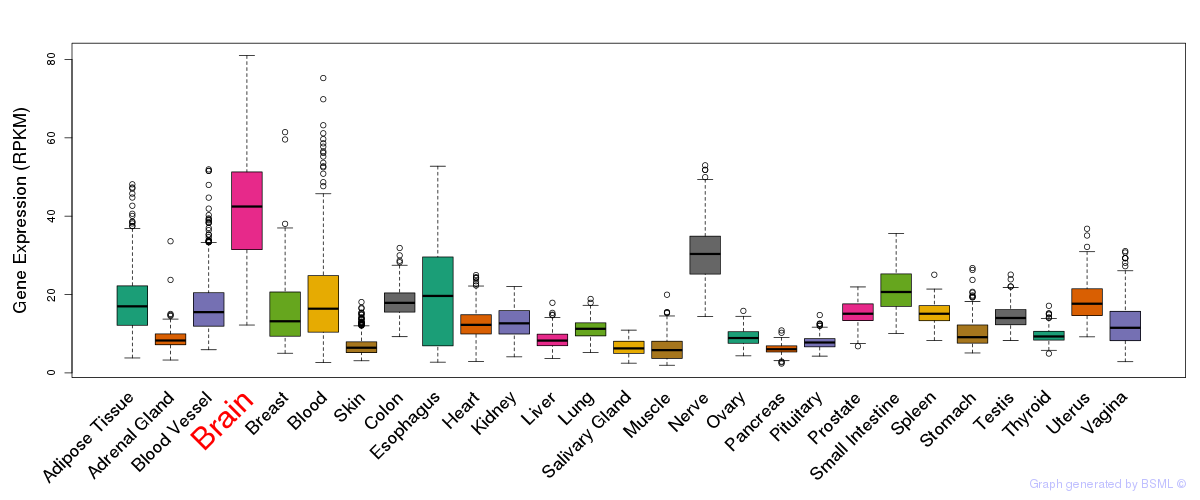
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PROX1 | 0.83 | 0.68 |
| FIGN | 0.80 | 0.66 |
| KITLG | 0.77 | 0.58 |
| ZIC2 | 0.76 | 0.44 |
| LHX9 | 0.76 | 0.27 |
| FZD10 | 0.76 | 0.44 |
| ZIC1 | 0.73 | 0.53 |
| KRT18P19 | 0.73 | 0.26 |
| TRPM8 | 0.72 | 0.49 |
| GBX2 | 0.72 | 0.36 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.22 | -0.24 |
| MT-CO2 | -0.22 | -0.24 |
| AC098691.2 | -0.22 | -0.32 |
| MYL3 | -0.21 | -0.18 |
| AF347015.33 | -0.20 | -0.18 |
| AF347015.8 | -0.20 | -0.20 |
| CA4 | -0.20 | -0.09 |
| AF347015.31 | -0.20 | -0.22 |
| AF347015.27 | -0.20 | -0.18 |
| AC018755.7 | -0.20 | -0.19 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG INOSITOL PHOSPHATE METABOLISM | 54 | 42 | All SZGR 2.0 genes in this pathway |
| KEGG PHOSPHATIDYLINOSITOL SIGNALING SYSTEM | 76 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | 132 | 101 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| LIU SOX4 TARGETS DN | 309 | 191 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL UP | 450 | 256 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR DN | 129 | 84 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| WANG BARRETTS ESOPHAGUS UP | 51 | 27 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| WILLIAMS ESR1 TARGETS UP | 26 | 16 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT REJECTED VS OK UP | 63 | 48 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C8 | 72 | 56 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| LIN NPAS4 TARGETS UP | 163 | 100 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC | 420 | 269 | All SZGR 2.0 genes in this pathway |
| ICHIBA GRAFT VERSUS HOST DISEASE D7 UP | 107 | 67 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 DN | 315 | 201 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF DN | 235 | 144 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 UP | 167 | 99 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| STEIN ESR1 TARGETS | 85 | 55 | All SZGR 2.0 genes in this pathway |
| STEIN ESTROGEN RESPONSE NOT VIA ESRRA | 18 | 12 | All SZGR 2.0 genes in this pathway |
| QI HYPOXIA | 140 | 96 | All SZGR 2.0 genes in this pathway |
| CHANGOLKAR H2AFY TARGETS UP | 48 | 28 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 7 | 403 | 240 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR BOUND ES | 462 | 273 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL DN | 308 | 187 | All SZGR 2.0 genes in this pathway |